Insulin-response epigenetic activation of Egr-1 and JunB genes at the nuclear periphery by A-type lamin-associated pY19-Caveolin-2 in the inner nuclear membrane
- PMID: 25753664
- PMCID: PMC4381080
- DOI: 10.1093/nar/gkv181
Insulin-response epigenetic activation of Egr-1 and JunB genes at the nuclear periphery by A-type lamin-associated pY19-Caveolin-2 in the inner nuclear membrane
Abstract
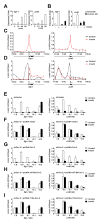
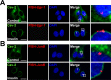
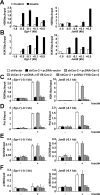
Insulin controls transcription to sustain its physiologic effects for the organism to adapt to environmental changes added to genetic predisposition. Nevertheless, insulin-induced transcriptional regulation by epigenetic factors and in defined nuclear territory remains elusive. Here we show that inner nuclear membrane (INM)-integrated caveolin-2 (Cav-2) regulates insulin-response epigenetic activation of Egr-1 and JunB genes at the nuclear periphery. INM-targeted pY19-Cav-2 in response to insulin associates specifically with the A-type lamin, disengages the repressed Egr-1 and JunB promoters from lamin A/C through disassembly of H3K9me3, and facilitates assembly of H3K9ac, H3K18ac and H3K27ac by recruitment of GCN5 and p300 and the subsequent enrichment of RNA polymerase II (Pol II) on the promoters at the nuclear periphery. Our findings show that Cav-2 is an epigenetic regulator of histone H3 modifications, and provide novel mechanisms of insulin-response epigenetic activation at the nuclear periphery.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- O'Brien R.M., Granner D.K. Regulation of gene expression by insulin. Physiol. Rev. 1996;76:1109–11061. - PubMed
-
- O'Brien R.M., Streeper R.S., Ayala J.E., Stadelmaier B.T., Hornbuckle L.A. Insulin-regulated gene expression. Biochem. Soc. Trans. 2001;29:552–558. - PubMed
-
- Rome S., Clément K., Rabasa-Lhoret R., Loizon E., Poitou C., Barsh G.S., Riou J.P., Laville M., Vidal H. Microarray profiling of human skeletal muscle reveals that insulin regulates approximately 800 genes during a hyperinsulinemic clamp. J. Biol. Chem. 2003;278:18063–18068. - PubMed
-
- Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. - PubMed
-
- Parton R.G., del Pozo M.A. Caveolae as plasma membrane sensors, protectors and organizers. Nat. Rev. Mol. Cell Biol. 2013;14:98–112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

