Dissecting the behavior and function of MBD3 in DNA methylation homeostasis by single-molecule spectroscopy and microscopy
- PMID: 25753672
- PMCID: PMC4381056
- DOI: 10.1093/nar/gkv098
Dissecting the behavior and function of MBD3 in DNA methylation homeostasis by single-molecule spectroscopy and microscopy
Abstract
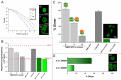
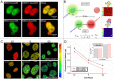
The detailed mechanism for DNA methylation homeostasis relies on an intricate regulatory network with a possible contribution from methyl-CpG-binding domain protein 3 (MBD3). In this study we examine the single-molecule behavior of MBD3 and its functional implication in balancing the activity of DNA methyltransferases (DNMTs). Besides a localization tendency to DNA demethylating sites, MBD3 experiences a concurrent transcription with DNMTs in cell cycle. Fluorescence lifetime correlation spectroscopy (FLCS) and photon counting histogram (PCH) were applied to characterize the chromatin binding kinetics and stoichiometry of MBD3 in different cell phases. In the G1-phase, MBD3, in the context of the Mi-2/NuRD (nucleosome remodeling deacetylase) complex, could adopt a salt-dependent homodimeric association with its target epigenomic loci. Along with cell cycle progression, utilizing fluorescence lifetime imaging microscopy-based Förster resonance energy transfer (FLIM-FRET) we revealed that a proportion of MBD3 and MBD2 would co-localize with DNMT1 during DNA maintenance methylation, providing a proofreading and protective mechanism against a possible excessive methylation by DNMT1. In accordance with our hypothesis, insufficient MBD3 induced by small interfering RNA (siRNA) was found to result in a global DNA hypermethylation as well as increased methylation in the promoter CpG islands (CGIs) of a number of cell cycle related genes.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

