Ubiquitination in the antiviral immune response
- PMID: 25753787
- PMCID: PMC4774549
- DOI: 10.1016/j.virol.2015.02.033
Ubiquitination in the antiviral immune response
Abstract
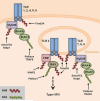
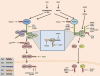
Ubiquitination has long been known to regulate fundamental cellular processes through the induction of proteasomal degradation of target proteins. More recently, 'atypical' non-degradative types of polyubiquitin chains have been appreciated as important regulatory moieties by modulating the activity or subcellular localization of key signaling proteins. Intriguingly, many of these non-degradative types of ubiquitination regulate the innate sensing pathways initiated by pattern recognition receptors (PRRs), ultimately coordinating an effective antiviral immune response. Here we discuss recent advances in understanding the functional roles of degradative and atypical types of ubiquitination in innate immunity to viral infections, with a specific focus on the signaling pathways triggered by RIG-I-like receptors, Toll-like receptors, and the intracellular viral DNA sensor cGAS.
Keywords: Antiviral immunity; E3 ligases; RIG-I-like receptors; STING; TRIM proteins; TRIM25; Toll-like receptors; Type-I interferon; Ubiquitin; cGAS.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



References
-
- Sen GC, Sarkar SN. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr Top Microbiol Immunol. 2007;316:23350. - PubMed
-
- Creagh EON. LA TLRs, NLRs and RLRs: A trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 2006;27:352–357. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

