Clinical implementation of integrated whole-genome copy number and mutation profiling for glioblastoma
- PMID: 25754088
- PMCID: PMC4578577
- DOI: 10.1093/neuonc/nov015
Clinical implementation of integrated whole-genome copy number and mutation profiling for glioblastoma
Abstract
Background: Multidimensional genotyping of formalin-fixed paraffin-embedded (FFPE) samples has the potential to improve diagnostics and clinical trials for brain tumors, but prospective use in the clinical setting is not yet routine. We report our experience with implementing a multiplexed copy number and mutation-testing program in a diagnostic laboratory certified by the Clinical Laboratory Improvement Amendments.
Methods: We collected and analyzed clinical testing results from whole-genome array comparative genomic hybridization (OncoCopy) of 420 brain tumors, including 148 glioblastomas. Mass spectrometry-based mutation genotyping (OncoMap, 471 mutations) was performed on 86 glioblastomas.
Results: OncoCopy was successful in 99% of samples for which sufficient DNA was obtained (n = 415). All clinically relevant loci for glioblastomas were detected, including amplifications (EGFR, PDGFRA, MET) and deletions (EGFRvIII, PTEN, 1p/19q). Glioblastoma patients ≤40 years old had distinct profiles compared with patients >40 years. OncoMap testing reliably identified mutations in IDH1, TP53, and PTEN. Seventy-seven glioblastoma patients enrolled on trials, of whom 51% participated in targeted therapeutic trials where multiplex data informed eligibility or outcomes. Data integration identified patients with complete tumor suppressor inactivation, albeit rarely (5% of patients) due to lack of whole-gene coverage in OncoMap.
Conclusions: Combined use of multiplexed copy number and mutation detection from FFPE samples in the clinical setting can efficiently replace singleton tests for clinical diagnosis and prognosis in most settings. Our results support incorporation of these assays into clinical trials as integral biomarkers and their potential to impact interpretation of results. Limited tumor suppressor variant capture by targeted genotyping highlights the need for whole-gene sequencing in glioblastoma.
Keywords: array CGH; clinical trials; genomics; genotyping; glioblastoma.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

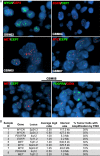
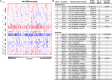

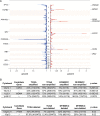
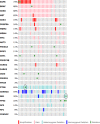
References
-
- Corless CL. Medicine. Personalized cancer diagnostics. Science. 2011;334(6060):1217–1218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

