The p53 isoform Δ133p53β promotes cancer stem cell potential
- PMID: 25754205
- PMCID: PMC4400643
- DOI: 10.1016/j.stemcr.2015.02.001
The p53 isoform Δ133p53β promotes cancer stem cell potential
Abstract
Cancer stem cells (CSC) are responsible for cancer chemoresistance and metastasis formation. Here we report that Δ133p53β, a TP53 splice variant, enhanced cancer cell stemness in MCF-7 breast cancer cells, while its depletion reduced it. Δ133p53β stimulated the expression of the key pluripotency factors SOX2, OCT3/4, and NANOG. Similarly, in highly metastatic breast cancer cells, aggressiveness was coupled with enhanced CSC potential and Δ133p53β expression. Like in MCF-7 cells, SOX2, OCT3/4, and NANOG expression were positively regulated by Δ133p53β in these cells. Finally, treatment of MCF-7 cells with etoposide, a cytotoxic anti-cancer drug, increased CSC formation and SOX2, OCT3/4, and NANOG expression via Δ133p53, thus potentially increasing the risk of cancer recurrence. Our findings show that Δ133p53β supports CSC potential. Moreover, they indicate that the TP53 gene, which is considered a major tumor suppressor gene, also acts as an oncogene via the Δ133p53β isoform.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

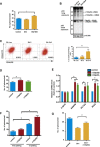
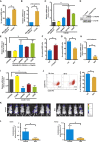
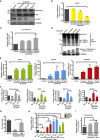
References
-
- Bernard H., Garmy-Susini B., Ainaoui N., Van Den Berghe L., Peurichard A., Javerzat S., Bikfalvi A., Lane D.P., Bourdon J.C., Prats A.C. The p53 isoform, Δ133p53α, stimulates angiogenesis and tumour progression. Oncogene. 2013;32:2150–2160. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

