Cutting, dicing, healing and sealing: the molecular surgery of tRNA
- PMID: 25755220
- PMCID: PMC4397177
- DOI: 10.1002/wrna.1279
Cutting, dicing, healing and sealing: the molecular surgery of tRNA
Abstract
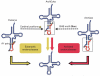
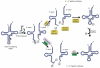
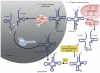
All organisms encode transfer RNAs (tRNAs) that are synthesized as precursor molecules bearing extra sequences at their 5' and 3' ends; some tRNAs also contain introns, which are removed by splicing. Despite commonality in what the ultimate goal is (i.e., producing a mature tRNA), mechanistically, tRNA splicing differs between Bacteria and Archaea or Eukarya. The number and position of tRNA introns varies between organisms and even between different tRNAs within the same organism, suggesting a degree of plasticity in both the evolution and persistence of modern tRNA splicing systems. Here we will review recent findings that not only highlight nuances in splicing pathways but also provide potential reasons for the maintenance of introns in tRNA. Recently, connections between defects in the components of the tRNA splicing machinery and medically relevant phenotypes in humans have been reported. These differences will be discussed in terms of the importance of splicing for tRNA function and in a broader context on how tRNA splicing defects can often have unpredictable consequences.
© 2015 John Wiley & Sons, Ltd.
Figures
References
-
- Fischer U, Englbrecht C, Chari A. Biogenesis of spliceosomal small nuclear ribonucleoproteins. WIREs RNA. 2011;2:718–731. - PubMed
-
- Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. - PubMed
-
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. - PubMed
-
- Kuhsel MG, Strickland R, Palmer JD. An ancient group I intron shared by eubacteria and chloroplasts. Science. 1990;250:1570–1573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

