An LXR-NCOA5 gene regulatory complex directs inflammatory crosstalk-dependent repression of macrophage cholesterol efflux
- PMID: 25755249
- PMCID: PMC4426483
- DOI: 10.15252/embj.201489819
An LXR-NCOA5 gene regulatory complex directs inflammatory crosstalk-dependent repression of macrophage cholesterol efflux
Abstract
LXR-cofactor complexes activate the gene expression program responsible for cholesterol efflux in macrophages. Inflammation antagonizes this program, resulting in foam cell formation and atherosclerosis; however, the molecular mechanisms underlying this antagonism remain to be fully elucidated. We use promoter enrichment-quantitative mass spectrometry (PE-QMS) to characterize the composition of gene regulatory complexes assembled at the promoter of the lipid transporter Abca1 following downregulation of its expression. We identify a subset of proteins that show LXR ligand- and binding-dependent association with the Abca1 promoter and demonstrate they differentially control Abca1 expression. We determine that NCOA5 is linked to inflammatory Toll-like receptor (TLR) signaling and establish that NCOA5 functions as an LXR corepressor to attenuate Abca1 expression. Importantly, TLR3-LXR signal crosstalk promotes recruitment of NCOA5 to the Abca1 promoter together with loss of RNA polymerase II and reduced cholesterol efflux. Together, these data significantly expand our knowledge of regulatory inputs impinging on the Abca1 promoter and indicate a central role for NCOA5 in mediating crosstalk between pro-inflammatory and anti-inflammatory pathways that results in repression of macrophage cholesterol efflux.
Keywords: LXR; NCOA5; atherosclerosis; inflammation; quantitative mass spectrometry.
© 2015 The Authors.
Figures
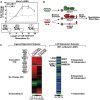
RT–qPCR of Abca1 transcripts following 1 μM T0901317 stimulation of LXR+/+ and LXR−/− primary BMMs. Fold changes are shown relative to LXR+/+ 0 h. Error bars represent ± SEM for n = 4–8 (**P = 0.001 at 8 h and **P = 0.00004 at 16 h versus LXR−/−; **P = 0.0003 at 0 h versus LXR+/+); bottom, proposed model explaining the ligand-dependent induction of Abca1. Following ligand stimulation, LXR–corepressor complexes are exchanged for LXR–coactivator complexes, which promote gene expression. The LXR–cofactor interactions responsible for the attenuation of expression beginning after 16 h remain unknown.
Overview of the PE-QMS experimental strategy to identify LXR ligand-stimulated and LXR binding-dependent interactions with the Abca1 promoter. Nuclear extracts were prepared from RAW 264.7 macrophages treated with control or 1 μM T0901317 (depicted as a black circle) for 18 h. The Abca1 promoter region was used to isolate gene regulatory complexes under the indicated conditions. Immobilized templates were washed, and bound proteins eluted and digested. Peptides were purified, then identified, and quantified by mass spectrometry.
Heat map of the PE-QMS experiment. Ligand-dependent subsets were quantified using label-free and isotope labeling approaches and are indicated on the left. Colors represent Log2 relative abundance ratios (T0901317/vehicle) of identified proteins. LXR binding-dependent subsets were quantified using an isotope labeling approach and are indicated on the right. Changes in binding > 1.5-fold were employed as cutoffs for depicted subsets. ND = not determined. Full dataset is available as Supplementary Table S1.

Protein–protein interaction network showing connectivity between the putative ligand-stimulated and LXR binding-dependent transcriptional regulators of Abca1 (shown in orange). Edges represent protein–protein interactions. NCOA5 node and edge are outlined red to indicate that this physical association is found in mouse interaction networks.
Luciferase reporter assays from RAW 264.7 macrophages expressing reporter alone, or together with full-length SND1, SART1, or HMBOX1. LXR ligand stimulations with 1 μM T0901317 were performed for 18 h. A diagram of the reporter construct is shown above. Fold changes are shown relative to vehicle-stimulated reporter alone (first lane). Error bars represent ± SEM for n = 9 (**P = 0.001, *P = 0.011 for SND1, *P = 0.046 for SART1).
Luciferase reporter assays from RAW 264.7 macrophages expressing reporter alone, or together with full-length NCOA5 or an NCOA5 mutant lacking the NH2-terminus. LXR ligand stimulations with 1 μM T0901317 were performed for 18 h. Diagrams of the reporter constructs are shown above. Fold changes are shown relative to vehicle-stimulated reporter alone (first lane). Error bars represent ± SEM for n = 6–12 (**P < 0.001, *P = 0.035).
RT–qPCR of Abca1 transcripts following infection of primary BMMs with Ncoa5 or control retrovirus. LXR ligand stimulations were performed with 1 μM T0901317 for 18 h. Fold changes are shown relative to vehicle-stimulated control. Error bars represent ± SEM for n = 4 (*P = 0.029).

In vitro pulldown assays using recombinant GST-LXRα to isolate the indicated in vitro translated NCOA5 constructs. Assays were performed for 2 h in the presence or absence of 2 μM T0901317. Samples were immunoblotted as indicated. Note the requirement of the NCOA5 NH2-terminus for interacting with LXRα.
Immunoprecipitation of Protein C-tagged LXRα from stable RAW 264.7 macrophage nuclear extracts stimulated with 1 μM T0901317 or vehicle control for 18 h, followed by immunoblotting for endogenous NCOA5 or over-expressed LXRα. Protein immunoblots of nuclear extracts are shown below.
NCOA5 ChIP time course from primary BMMs stimulated with 1 μM T0901317 or vehicle control. qPCR was performed for the Abca1 proximal LXRE. Note the ligand-stimulated association of NCOA5. Error bars represent ± SEM for n = 4–9 (*P = 0.02).
NCOA5 ChIP assays from LXR−/− BMMs stimulated with 1 μM T0901317 or vehicle control for 18 h. qPCR was performed for the Abca1 proximal LXRE. Error bars represent ± SEM for n = 5–6 (**P = 0.0003 for vehicle and **P = 0.002 for T0901317).

RT–qPCR of Abca1 expression following 1 μM T0901317 treatment in primary BMMs infected with a non-silencing or Ncoa5-specific shRNA. Fold changes are shown relative to shControl 0 h. Note the elevated Abca1 expression at 24–30 h following loss of NCOA5. Error bars represent ± SEM for n = 4–6 (**P = 0.0007 at 24 h and **P = 0.004 at 30 h, *P = 0.02).
Quantification of ABCA1 immunoblots from primary BMMs infected as in (A). Representative immunoblot is shown in Supplementary Fig S7E. LXR ligand stimulation with 1 μM T0901317 or vehicle control was performed for 8 h or 30 h. Error bars represent ± SEM for n = 2 (**P = 0.007).

A, B RT–qPCR of Abca1 expression from primary BMMs infected with non-silencing or Ncoa5-specific shRNAs. Ligand stimulations were performed for 4 h with vehicle control, 1 μM T0901317 alone or together with 6 μg/ml PolyIC (A) or 10 ng/ml LPS (B). Fold changes are shown relative to vehicle-stimulated shControl. Note only the loss of TLR3-mediated repression following Ncoa5 silencing. Error bars represent ± SEM for n = 4–10 (**P = 0.0003 for A, **P = 0.0004 for shControl in B, **P = 0.0002 for shNcoa5 in B versus T0901317).
C, D RT–qPCR of Abca1 expression from primary BMMs infected with non-silencing or Ncoa5-specific shRNAs. Ligand stimulations were performed for 4 h unstimulated, or with 6 μg/ml PolyIC (C) or 10 ng/ml LPS (D). Fold changes are shown relative to unstimulated shControl. Error bars represent ± SEM for n = 4–6 (**P = 0.0006 for C, **P = 0.00000001 for shControl in D, **P = 0.000001 for shNcoa5 in D, *P = 0.013 versus T0901317).
E NCOA5 ChIP assays from primary BMMs stimulated for 3 h with 1 μM T0901317, 1 μM T0901317 + 6 μg/ml PolyIC, or vehicle control. qPCR was performed for the Abca1 proximal LXRE. Note the increased occupancy only in the presence of both lipid and inflammatory ligands. Error bars represent ± SEM for n = 8–12 (**P = 0.0007 versus T0901317).
F NCOA5 ChIP assays from primary BMMs stimulated for 3 h with 6 μg/ml PolyIC or left unstimulated. qPCR was performed for the Abca1 proximal LXRE. Error bars represent ± SEM for n = 5–6.
G Cholesterol efflux assays to APOA1 from primary BMMs infected with non-silencing or Ncoa5-specific shRNAs. Agonist stimulations were performed for 6 h. Error bars represent ± SEM for n = 3 (*P = 0.02 versus T0901317).

A, B Unmodified/pSer5 RNAPII (A) or RNAPII pSer2 (B) ChIP assays from primary BMMs stimulated for 4 h with 1 μM T0901317, 1 μM T0901317 + 6 μg/ml PolyIC, or vehicle control. qPCR was performed for the Abca1 TSS. Error bars represent ± SEM for n = 6–9 (**P = 0.0001 in A, **P = 0.002 in B versus T0901317).
C, D RNAPII ChIP assays as in (A, B) but performed from LXR−/− BMMs. Error bars represent ± SEM for n = 5–9 (**P = 0.000000001, *P = 0.02 versus T0901317).
E RT–qPCR of Abca1 expression from LXR+/+ versus LXR−/− BMMs. Ligand stimulations were performed for 4 h with vehicle control, 1 μM T0901317, or 1 μM T0901317 together with 6 μg/ml PolyIC. Fold changes are shown relative to vehicle-stimulated LXR+/+. Error bars represent ± SEM for n = 4 (**P = 0.004, *P = 0.012 versus T0901317).
F RNAPII pSer2 ChIP assays from primary BMMs infected with non-silencing or Ncoa5-specific shRNAs. Ligand stimulations were performed for 4 h with vehicle control, 1 μM T0901317, or 1 μM T0901317 + 6 μg/ml PolyIC. qPCR was performed for the Abca1 TSS. Note RNAPII pSer2 returns to baseline occupancy following TLR3 stimulation in shControl but not shNcoa5 BMMs (**P = 0.004 versus baseline). Error bars represent ± SEM for n = 3 from 11 mice.
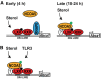
In response to sterol ligand treatment, LXR–RXR heterodimers initially induce the transcription of Abca1 through recruitment and activation of RNAPII (left). Following prolonged sterol ligand treatment, the NCOA5 repressor directly interacts with LXR at the Abca1 promoter. This repressor complex inhibits the recruitment and function of RNAPII resulting in attenuated expression of Abca1 (right). We hypothesize that prolonged sterol ligand treatment induces a regulatory modification on NCOA5 (shown in green), such as phosphorylation, which facilitates its recruitment to LXR.
Crosstalk between pro-inflammatory TLR3 and anti-inflammatory LXR pathways promotes the association of NCOA5 with LXR, resulting in the inhibition of RNAPII recruitment and function, and repression of Abca1 gene expression. The regulatory modification of NCOA5 following activation of these two pathways may be similar or distinct to that in (A). We also cannot discount the contribution of additional constituents of this transcriptional complex to the recruitment of NCOA5.
Similar articles
-
DNA topoisomerase II inhibitors induce macrophage ABCA1 expression and cholesterol efflux-an LXR-dependent mechanism.Biochim Biophys Acta. 2013 Jun;1831(6):1134-45. doi: 10.1016/j.bbalip.2013.02.007. Epub 2013 Mar 1. Biochim Biophys Acta. 2013. PMID: 23466610
-
Poly(ADP-ribose) Polymerase 1 Represses Liver X Receptor-mediated ABCA1 Expression and Cholesterol Efflux in Macrophages.J Biol Chem. 2016 May 20;291(21):11172-84. doi: 10.1074/jbc.M116.726729. Epub 2016 Mar 29. J Biol Chem. 2016. PMID: 27026705 Free PMC article.
-
Crosstalk between LXR and toll-like receptor signaling mediates bacterial and viral antagonism of cholesterol metabolism.Mol Cell. 2003 Oct;12(4):805-16. doi: 10.1016/s1097-2765(03)00384-8. Mol Cell. 2003. PMID: 14580333
-
The crosstalk of ABCA1 and ANXA1: a potential mechanism for protection against atherosclerosis.Mol Med. 2020 Sep 7;26(1):84. doi: 10.1186/s10020-020-00213-y. Mol Med. 2020. PMID: 32894039 Free PMC article. Review.
-
PPARgamma1 and LXRalpha face a new regulator of macrophage cholesterol homeostasis and inflammatory responsiveness, AEBP1.Nucl Recept Signal. 2010 Apr 16;8:e004. doi: 10.1621/nrs.08004. Nucl Recept Signal. 2010. PMID: 20419060 Free PMC article. Review.
Cited by
-
Proteomic analysis identifies transcriptional cofactors and homeobox transcription factors as TBX18 binding proteins.PLoS One. 2018 Aug 2;13(8):e0200964. doi: 10.1371/journal.pone.0200964. eCollection 2018. PLoS One. 2018. PMID: 30071041 Free PMC article.
-
A novel tumor suppressor gene NCOA5 is correlated with progression in papillary thyroid carcinoma.Onco Targets Ther. 2018 Jan 11;11:307-311. doi: 10.2147/OTT.S154158. eCollection 2018. Onco Targets Ther. 2018. PMID: 29391807 Free PMC article.
-
C10orf99/GPR15L Regulates Proinflammatory Response of Keratinocytes and Barrier Formation of the Skin.Front Immunol. 2022 Feb 22;13:825032. doi: 10.3389/fimmu.2022.825032. eCollection 2022. Front Immunol. 2022. PMID: 35273606 Free PMC article.
-
NCOA5 Haplo-insufficiency Results in Male Mouse Infertility through Increased IL-6 Expression in the Epididymis.Sci Rep. 2019 Oct 29;9(1):15525. doi: 10.1038/s41598-019-52105-9. Sci Rep. 2019. PMID: 31664153 Free PMC article.
-
NCOA5 promotes proliferation, migration and invasion of colorectal cancer cells via activation of PI3K/AKT pathway.Oncotarget. 2017 Nov 14;8(64):107932-107946. doi: 10.18632/oncotarget.22429. eCollection 2017 Dec 8. Oncotarget. 2017. PMID: 29296214 Free PMC article.
References
-
- Ahmad U, Ali R, Lebastchi AH, Qin L, Lo SF, Yakimov AO, Khan SF, Choy JC, Geirsson A, Pober JS, Tellides G. IFN-gamma primes intact human coronary arteries and cultured coronary smooth muscle cells to double-stranded RNA- and self-RNA-induced inflammatory responses by upregulating TLR3 and melanoma differentiation-associated gene 5. J Immunol. 2010;185:1283–1294. - PMC - PubMed
-
- Aiello RJ, Brees D, Bourassa PA, Royer L, Lindsey S, Coskran T, Haghpassand M, Francone OL. Increased atherosclerosis in hyperlipidemic mice with inactivation of ABCA1 in macrophages. Arterioscler Thromb Vasc Biol. 2002;22:630–637. - PubMed
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. - PMC - PubMed
-
- Baiersdorfer M, Schwarz M, Seehafer K, Lehmann C, Heit A, Wagner H, Kirschning CJ, Koch-Brandt C. Toll-like receptor 3 mediates expression of clusterin/apolipoprotein J in vascular smooth muscle cells stimulated with RNA released from necrotic cells. Exp Cell Res. 2010;316:3489–3500. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

