Molecular biology of the hepatitis B virus for clinicians
- PMID: 25755457
- PMCID: PMC3940099
- DOI: 10.1016/j.jceh.2012.10.003
Molecular biology of the hepatitis B virus for clinicians
Abstract
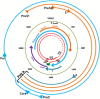
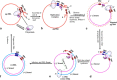
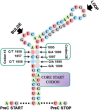
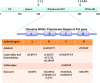
Hepatitis B virus (HBV) infection is one of the major global health problems, especially in economically under-developed or developing countries. HBV infection can lead to a number of clinical outcomes including chronic infection, cirrhosis and liver cancer. It ranks among the top 10 causes of death, being responsible for around 1 million deaths every year. Despite the availability of a highly efficient vaccine and potent antiviral agents, HBV infection still remains a significant clinical problem, particularly in those high endemicity areas where vaccination of large populations has not been possible due to economic reasons. Although HBV is among the smallest viruses in terms of virion and genome size, it has numerous unique features that make it completely distinct from other DNA viruses. It has a partially double stranded DNA with highly complex genome organization, life cycle and natural history. Remarkably distinct from other DNA viruses, it uses an RNA intermediate called pregenomic RNA (pgRNA) and reverse transcriptase for its genome replication. Genome replication is accomplished by a complex mechanism of primer shifting facilitated by direct repeat sequences encoded in the genome. Further, the genome has evolved in such a manner that every single nucleotide of the genome is used for either coding viral proteins or used as regulatory regions or both. Moreover, it utilizes internal in-frame translation initiation codons, as well as different reading frames from the same RNA to generate different proteins with diverse functions. HBV also shows considerable genetic variability which has been related with clinical outcomes, replication potential, therapeutic response etc. This review aims at reviewing fundamental events of the viral life cycle including viral replication, transcription and translation, from the molecular standpoint, as well as, highlights the clinical relevance of genetic variability of HBV.
Keywords: AUG, translation start codon; BCP, basal core promoter; CHB, chronic hepatitis B infection; DR, direct repeat; EBP, enhancer binding protein; EN, enhancer; ER, endoplasmic reticulum; HBV, hepatitis B virus; HBsAg; HCC, hepatocellular cancer; Hepadnavirus; IL, interleukin; LEF, liver enriched factors; LHB, large envelope protein; MHBs, middle hepatitis B surface antigen; MHR, major hydrophilic region; ORF, open reading frames; PC, precore; RT, reverse transcriptase; SHBs, small hepatitis B surface antigen; TGF-α, transforming growth factor-α; TNF-α, tumor necrosis factor-α; TP, terminal protein; WHV, woodchuck hepatitis virus; cccDNA, covalently closed circular; dGMP, deoxyguanosine monophosphate; genotype; pHSA, poly-human serum albumin; pgRNA; pgRNA, pregenomic RNA; rcDNA; rcDNA, relaxed circular DNA.
Figures


References
-
- European Association for the Study of the Liver EASL Clinical Practice Guidelines: management of chronic hepatitis B. J Hepatol. 2012;57:167–185. - PubMed
-
- Beasley R.P., Hwang L.Y., Lin C.C., Chien C.S. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22,707 men in Taiwan. Lancet. 1981;2:1129–1133. - PubMed
-
- Shepard C.W., Simard E.P., Finelli L., Fiore A.E., Bell B.P. Hepatitis B virus infection: epidemiology and vaccination. Epidemiol Rev. 2006;28:112–125. - PubMed
-
- Lurman A. Eine icterus epidemic. Berl Klin Woschenschr. 1885;22:20–23. [in German]
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

