Detection of MGMT promoter methylation in glioblastoma using pyrosequencing
- PMID: 25755756
- PMCID: PMC4348884
Detection of MGMT promoter methylation in glioblastoma using pyrosequencing
Abstract
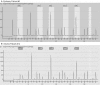
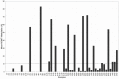
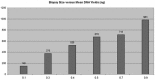
Recent clinical trials on patients with glioblastoma revealed that O(6)-Methylguanine-DNA methyltransferase (MGMT) methylation status significantly predicts patient's response to alkylating agents. In this study, we sought to develop and validate a quantitative MGMT methylation assay using pyrosequencing on glioblastoma. We quantified promoter methylation of MGMT using pyrosequencing on paraffin-embedded fine needle aspiration biopsy tissues from 43 glioblastoma. Using a 10% cutoff, MGMT methylation was identified in 37% cases of glioblastoma and 0% of the non-neoplastic epileptic tissue. Methylation of any individual CpG island in MGMT promoter ranged between 33% and 95%, with a mean of 65%. By a serial dilution of genomic DNA of a homogenously methylated cancer cell line with an unmethylated cell line, the analytical sensitivity is at 5% for pyrosequencing to detect MGMT methylation. The minimal amount of genomic DNA required is 100 ng (approximately 3,000 cells) in small fine needle biopsy specimens. Compared with methylation-specific PCR, pyrosequencing is comparably sensitive, relatively specific, and also provides quantitative information for each CpG methylation.
Keywords: MGMT; epigenetics; glioblastoma; methylation; pyrosequencing.
Figures
References
-
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. - PubMed
-
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–4. - PubMed
-
- Karayan-Tapon L, Quillien V, Guilhot J, Wager M, Fromont G, Saikali S, Etcheverry A, Hamlat A, Loussouarn D, Campion L, Campone M, Vallette FM, Gratas-Rabbia-Ré C. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2010;97:311–22. - PubMed
-
- Esteller M, Herman JG. Generating mutations but providing chemosensitivity: the role of O6-methylguanine DNA methyltransferase in human cancer. Oncogene. 2004;23:1–8. - PubMed
-
- Paz MF, Yaya-Tur R, Rojas-Marcos I, Reynes G, Pollan M, Aguirre-Cruz L, García-Lopez JL, Piquer J, Safont MJ, Balaña C, Sanchez-Cespedes M, García-Villanueva M, Arribas L, Esteller M. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res. 2004;10:4933–8. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
