Differentiation between acute skin rejection in allotransplantation and T-cell mediated skin inflammation based on gene expression analysis
- PMID: 25756043
- PMCID: PMC4338383
- DOI: 10.1155/2015/259160
Differentiation between acute skin rejection in allotransplantation and T-cell mediated skin inflammation based on gene expression analysis
Abstract
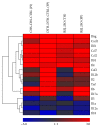
Advances in microsurgical techniques and immunosuppressive medication have rendered transplantation of vascularized composite allografts possible, when autologous tissue is neither available nor sufficient for reconstruction. However, skin rejection and side effects of long-term immunosuppression still remain a major hurdle for wide adoption of this excellent reconstructive technique. Histopathologic changes during acute skin rejection in vascular composite allotransplantation often mimic inflammatory skin disorders and are hard to distinguish. Hence, the identification of diagnostic and therapeutic markers specific for skin rejection is of particular clinical need. Here we present novel markers allowing for early differentiation between rejection in hind limb allotransplantation and contact hypersensitivity. Assessment of Ccl7, Il18, and Il1b expression is most indicative of distinguishing skin rejection from skin inflammatory disorders. Gene expression levels varied significantly across skin types and regions, indicating localization specific mechanism of leukocyte migration and infiltration. Expression of Il12b, Il17a, and Il1b gene expression levels differed significantly between rejection and inflammation, independent of the skin type. In synopsis of the RNA expression profile and previously assessed protein expression, the Il1 family appears as a promising option for accurate skin rejection diagnosis and, as a following step, for development of novel rejection treatments.
Figures





References
-
- Saint-Mezard P., Berard F., Dubois B., Kaiserlian D., Nicolas J. F. The role of CD4+ and CD8+ T cells in contact hypersensitivity and allergic contact dermatitis. European Journal of Dermatology. 2004;14(3):131–138. - PubMed
-
- Pastore S., Mascia F., Mariotti F., Dattilo C., Girolomoni G. Chemokine networks in inflammatory skin diseases. European Journal of Dermatology. 2004;14(4):203–208. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

