Overexpression of an outer membrane protein associated with decreased susceptibility to carbapenems in Proteus mirabilis
- PMID: 25756370
- PMCID: PMC4355480
- DOI: 10.1371/journal.pone.0120395
Overexpression of an outer membrane protein associated with decreased susceptibility to carbapenems in Proteus mirabilis
Abstract
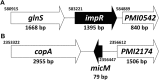
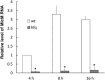
Proteus mirabilis isolates commonly have decreased susceptibility to imipenem. Previously, we found P. mirabilis hfq mutant was more resistant to imipenem and an outer membrane protein (OMP) could be involved. Therefore, we investigated the role of this OMP in carbapenem susceptibility. By SDS-PAGE we found this OMP (named ImpR) was increased in hfq mutant and LC-MS/MS revealed it to be the homologue of Salmonella YbfM, which is a porin for chitobiose and subject to MicM (a small RNA) regulation. We demonstrated that ImpR overexpression resulted in increased carbapenem MICs in the laboratory strain and clinical isolates. Chitobiose induced expression of chb (a chitobiose utilization operon). Real-time RT-PCR and SDS-PAGE were performed to elucidate the relationship of hfq, impR, chb and MicM in P. mirabilis. We found MicM RNA was decreased in hfq mutant and chbBC-intergenic region (chbBC-IGR) overexpression strain (chbIGRov), while impR mRNA was increased in hfq mutant, micM mutant and chbIGRov strain. In addition, mutation of hfq or micM and overexpression of chbBC-IGR increased ImpR protein level. Accordingly, chitobiose made wild-type have higher levels of ImpR protein and are more resistant to carbapenems. Hfq- and MicM-complemented strains restored wild-type MICs. Mutation of both impR and hfq eliminated the increase in carbapenem MICs observed in hfq mutant and ImpR-complementation of hfq/impR double mutant resulted in MICs as hfq mutant, indicating that the ImpR-dependent decreased carbapenem susceptibility of hfq mutant. These indicate MicM was antisense to impR mRNA and was negatively-regulated by chbBC-IGR. Together, overexpression of ImpR contributed to the decreased carbapenem susceptibility in P. mirabilis.
Conflict of interest statement
Figures




References
-
- Jiang SS, Lin TY, Wang WB, Liu MC, Hsueh PR, Liaw SJ. Characterization of UDP-glucose dehydrogenase and UDP-glucose pyrophosphorylase mutants of Proteus mirabilis: defectiveness in polymyxin B resistance, swarming, and virulence. Antimicrob Agents Chemother. 2010;54: 2000–2009. 10.1128/AAC.01384-09 - DOI - PMC - PubMed
-
- Tibbetts R, Frye JG, Marschall J, Warren D, Dunne W. Detection of KPC-2 in a clinical isolate of Proteus mirabilis and first reported description of carbapenemase resistance caused by a KPC beta-lactamase in P. mirabilis . J Clin Microbiol. 2008;46: 3080–3083. 10.1128/JCM.00979-08 - DOI - PMC - PubMed
-
- Tenover FC, Emery SL, Spiegel CA, Bradford PA, Eells S, Endimiani A, et al. Identification of plasmid-mediated AmpC beta-lactamases in Escherichia coli, Klebsiella spp., and Proteus species can potentially improve reporting of cephalosporin susceptibility testing results. J Clin Microbiol. 2009;47: 294–299. 10.1128/JCM.01797-08 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

