The evolutionary and functional diversity of classical and lesser-known cytoplasmic and organellar translational GTPases across the tree of life
- PMID: 25756599
- PMCID: PMC4342817
- DOI: 10.1186/s12864-015-1289-7
The evolutionary and functional diversity of classical and lesser-known cytoplasmic and organellar translational GTPases across the tree of life
Abstract
Background: The ribosome translates mRNA to protein with the aid of a number of accessory protein factors. Translational GTPases (trGTPases) are an integral part of the 'core set' of essential translational factors, and are some of the most conserved proteins across life. This study takes advantage of the wealth of available genomic data, along with novel functional information that has come to light for a number of trGTPases to address the full evolutionary and functional diversity of this superfamily across all domains of life.
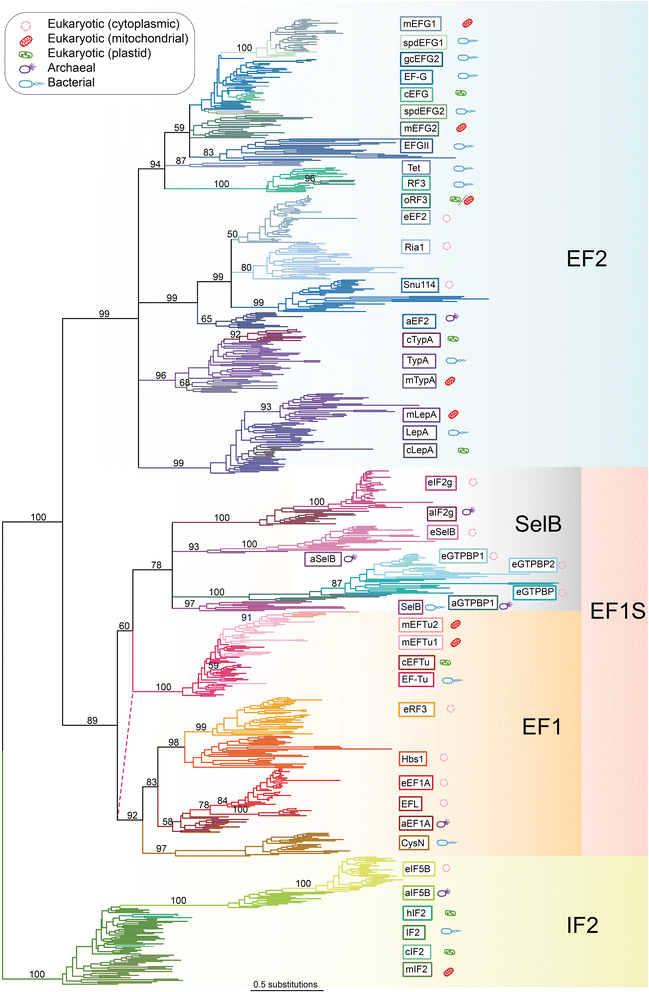
Results: Through sensitive sequence searching combined with phylogenetic analysis, 57 distinct subfamilies of trGTPases are identified: 14 bacterial, 7 archaeal and 35 eukaryotic (of which 21 are known or predicted to be organellar). The results uncover the functional evolution of trGTPases from before the last common ancestor of life on earth to the current day.
Conclusions: While some trGTPases are universal, others are limited to certain taxa, suggesting lineage-specific translational control mechanisms that exist on a base of core factors. These lineage-specific features may give organisms the ability to tune their translation machinery to respond to their environment. Only a fraction of the diversity of the trGTPase superfamily has been subjected to experimental analyses; this comprehensive classification brings to light novel and overlooked translation factors that are worthy of further investigation.
Figures

References
-
- Voorhees RM, Ramakrishnan V. Structural basis of the translational elongation cycle. Annu Rev Biochem. 2013;82:203–36. - PubMed
-
- Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19(6):568–76. - PubMed
-
- Myasnikov AG, Simonetti A, Marzi S, Klaholz BP. Structure-function insights into prokaryotic and eukaryotic translation initiation. Curr Opin Struct Biol. 2009;19(3):300–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

