Adaptive Evolution as a Predictor of Species-Specific Innate Immune Response
- PMID: 25758009
- PMCID: PMC4476151
- DOI: 10.1093/molbev/msv051
Adaptive Evolution as a Predictor of Species-Specific Innate Immune Response
Abstract
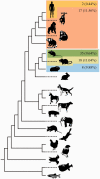
It has been proposed that positive selection may be associated with protein functional change. For example, human and macaque have different outcomes to HIV infection and it has been shown that residues under positive selection in the macaque TRIM5α receptor locate to the region known to influence species-specific response to HIV. In general, however, the relationship between sequence and function has proven difficult to fully elucidate, and it is the role of large-scale studies to help bridge this gap in our understanding by revealing major patterns in the data that correlate genotype with function or phenotype. In this study, we investigate the level of species-specific positive selection in innate immune genes from human and mouse. In total, we analyzed 456 innate immune genes using codon-based models of evolution, comparing human, mouse, and 19 other vertebrate species to identify putative species-specific positive selection. Then we used population genomic data from the recently completed Neanderthal genome project, the 1000 human genomes project, and the 17 laboratory mouse genomes project to determine whether the residues that were putatively positively selected are fixed or variable in these populations. We find evidence of species-specific positive selection on both the human and the mouse branches and we show that the classes of genes under positive selection cluster by function and by interaction. Data from this study provide us with targets to test the relationship between positive selection and protein function and ultimately to test the relationship between positive selection and discordant phenotypes.
Keywords: adaptive evolution; innate immune evolution; predicting phenotypic response; protein functional shift; species-specific responses.
© The Author 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

