Dynamic visualization of transcription and RNA subcellular localization in zebrafish
- PMID: 25758462
- PMCID: PMC4378251
- DOI: 10.1242/dev.118968
Dynamic visualization of transcription and RNA subcellular localization in zebrafish
Abstract
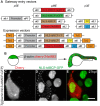
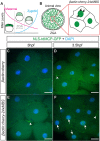
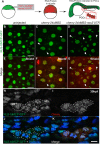
Live imaging of transcription and RNA dynamics has been successful in cultured cells and tissues of vertebrates but is challenging to accomplish in vivo. The zebrafish offers important advantages to study these processes--optical transparency during embryogenesis, genetic tractability and rapid development. Therefore, to study transcription and RNA dynamics in an intact vertebrate organism, we have adapted the MS2 RNA-labeling system to zebrafish. By using this binary system to coexpress a fluorescent MS2 bacteriophage coat protein (MCP) and an RNA of interest tagged with multiple copies of the RNA hairpin MS2-binding site (MBS), live-cell imaging of RNA dynamics at single RNA molecule resolution has been achieved in other organisms. Here, using a Gateway-compatible MS2 labeling system, we generated stable transgenic zebrafish lines expressing MCP, validated the MBS-MCP interaction and applied the system to investigate zygotic genome activation (ZGA) and RNA localization in primordial germ cells (PGCs) in zebrafish. Although cleavage stage cells are initially transcriptionally silent, we detect transcription of MS2-tagged transcripts driven by the βactin promoter at ∼ 3-3.5 h post-fertilization, consistent with the previously reported ZGA. Furthermore, we show that MS2-tagged nanos3 3'UTR transcripts localize to PGCs, where they are diffusely cytoplasmic and within larger cytoplasmic accumulations reminiscent of those displayed by endogenous nanos3. These tools provide a new avenue for live-cell imaging of RNA molecules in an intact vertebrate. Together with new techniques for targeted genome editing, this system will be a valuable tool to tag and study the dynamics of endogenous RNAs during zebrafish developmental processes.
Keywords: In vivo RNA labeling; MS2; Transcription; Transgenic zebrafish.
© 2015. Published by The Company of Biologists Ltd.
Figures

Similar articles
-
Visualization of Turbot (Scophthalmus maximus) Primordial Germ Cells in vivo Using Fluorescent Protein Mediated by the 3' Untranslated Region of nanos3 or vasa Gene.Mar Biotechnol (NY). 2019 Oct;21(5):671-682. doi: 10.1007/s10126-019-09911-z. Epub 2019 Sep 9. Mar Biotechnol (NY). 2019. PMID: 31502176
-
Visualization of primordial germ cells in the fertilized pelagic eggs of the barfin flounder Verasper moseri.Int J Dev Biol. 2015;59(10-12):465-70. doi: 10.1387/ijdb.150008rg. Int J Dev Biol. 2015. PMID: 26864487
-
Visualization of primordial germ cells in vivo using GFP-nos1 3'UTR mRNA.Int J Dev Biol. 2006;50(8):691-9. doi: 10.1387/ijdb.062143ts. Int J Dev Biol. 2006. PMID: 17051479
-
Genetic tools for multicolor imaging in zebrafish larvae.Methods. 2013 Aug 15;62(3):279-91. doi: 10.1016/j.ymeth.2013.07.028. Epub 2013 Jul 22. Methods. 2013. PMID: 23886907 Review.
-
Transgenic fish systems and their application in ecotoxicology.Crit Rev Toxicol. 2015 Feb;45(2):124-41. doi: 10.3109/10408444.2014.965805. Epub 2014 Nov 14. Crit Rev Toxicol. 2015. PMID: 25394772 Review.
Cited by
-
Imaging nascent transcription in wholemount vertebrate embryos to characterize zygotic genome activation.Methods Enzymol. 2020;638:139-165. doi: 10.1016/bs.mie.2020.03.002. Epub 2020 Mar 23. Methods Enzymol. 2020. PMID: 32416911 Free PMC article.
-
A simple MiMIC-based approach for tagging endogenous genes to visualise live transcription in Drosophila.Development. 2024 Dec 15;151(24):dev204294. doi: 10.1242/dev.204294. Epub 2024 Dec 16. Development. 2024. PMID: 39584418 Free PMC article.
-
The Akt-mTOR Pathway Drives Myelin Sheath Growth by Regulating Cap-Dependent Translation.J Neurosci. 2021 Oct 13;41(41):8532-8544. doi: 10.1523/JNEUROSCI.0783-21.2021. Epub 2021 Sep 2. J Neurosci. 2021. PMID: 34475201 Free PMC article.
-
mRNA quantification using single-molecule FISH in Drosophila embryos.Nat Protoc. 2017 Jul;12(7):1326-1348. doi: 10.1038/nprot.2017.030. Epub 2016 Jun 8. Nat Protoc. 2017. PMID: 28594816 Free PMC article.
-
A cell cycle-coordinated Polymerase II transcription compartment encompasses gene expression before global genome activation.Nat Commun. 2019 Feb 11;10(1):691. doi: 10.1038/s41467-019-08487-5. Nat Commun. 2019. PMID: 30741925 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- EB013571/EB/NIBIB NIH HHS/United States
- P30CA013330/CA/NCI NIH HHS/United States
- R01GM089979/GM/NIGMS NIH HHS/United States
- R01 GM057071/GM/NIGMS NIH HHS/United States
- NS083085/NS/NINDS NIH HHS/United States
- F31 NS083258/NS/NINDS NIH HHS/United States
- R01 NS083085/NS/NINDS NIH HHS/United States
- R01GM057071/GM/NIGMS NIH HHS/United States
- R01 GM089979/GM/NIGMS NIH HHS/United States
- R01 EB013571/EB/NIBIB NIH HHS/United States
- T32 GM007288/GM/NIGMS NIH HHS/United States
- 1F31NS083258/NS/NINDS NIH HHS/United States
- P30 CA013330/CA/NCI NIH HHS/United States
- T32-GM007288/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

