Involvement of a citrus meiotic recombination TTC-repeat motif in the formation of gross deletions generated by ionizing radiation and MULE activation
- PMID: 25758634
- PMCID: PMC4334395
- DOI: 10.1186/s12864-015-1280-3
Involvement of a citrus meiotic recombination TTC-repeat motif in the formation of gross deletions generated by ionizing radiation and MULE activation
Abstract
Background: Transposable-element mediated chromosomal rearrangements require the involvement of two transposons and two double-strand breaks (DSB) located in close proximity. In radiobiology, DSB proximity is also a major factor contributing to rearrangements. However, the whole issue of DSB proximity remains virtually unexplored.
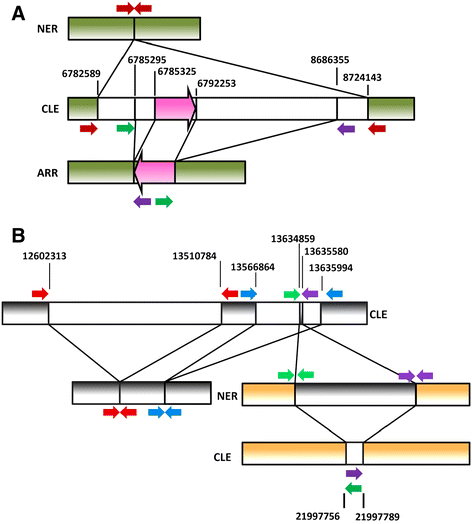
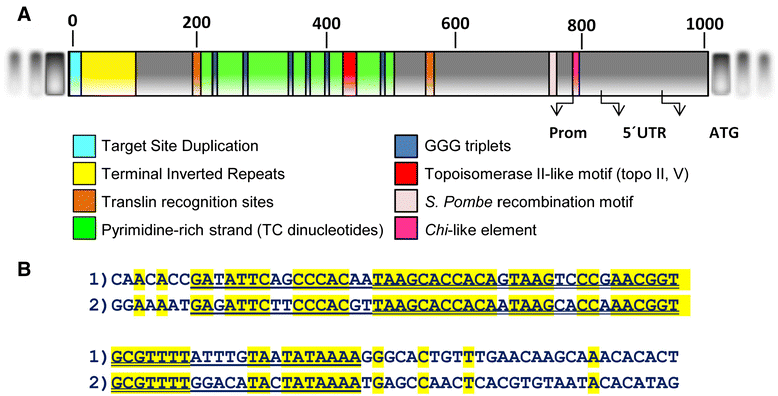
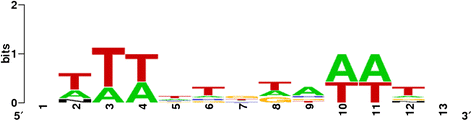
Results: Based on DNA sequencing analysis we show that the genomes of 2 derived mutations, Arrufatina (sport) and Nero (irradiation), share a similar 2 Mb deletion of chromosome 3. A 7 kb Mutator-like element found in Clemenules was present in Arrufatina in inverted orientation flanking the 5' end of the deletion. The Arrufatina Mule displayed "dissimilar" 9-bp target site duplications separated by 2 Mb. Fine-scale single nucleotide variant analyses of the deleted fragments identified a TTC-repeat sequence motif located in the center of the deletion responsible of a meiotic crossover detected in the citrus reference genome.
Conclusions: Taken together, this information is compatible with the proposal that in both mutants, the TTC-repeat motif formed a triplex DNA structure generating a loop that brought in close proximity the originally distinct reactive ends. In Arrufatina, the loop brought the Mule ends nearby the 2 distinct insertion target sites and the inverted insertion of the transposable element between these target sites provoked the release of the in-between fragment. This proposal requires the involvement of a unique transposon and sheds light on the unresolved question of how two distinct sites become located in close proximity. These observations confer a crucial role to the TTC-repeats in fundamental plant processes as meiotic recombination and chromosomal rearrangements.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

