The purification and characterization of ATP synthase complexes from the mitochondria of four fungal species
- PMID: 25759169
- PMCID: PMC4422255
- DOI: 10.1042/BJ20150197
The purification and characterization of ATP synthase complexes from the mitochondria of four fungal species
Abstract
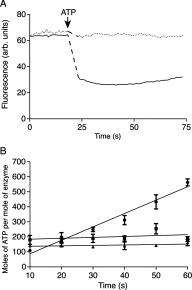
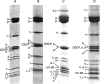
The ATP synthases have been isolated by affinity chromatography from the mitochondria of the fungal species Yarrowia lipolytica, Pichia pastoris, Pichia angusta and Saccharomyces cerevisiae. The subunit compositions of the purified enzyme complexes depended on the detergent used to solubilize and purify the complex, and the presence or absence of exogenous phospholipids. All four enzymes purified in the presence of n-dodecyl-β-D-maltoside had a complete complement of core subunits involved directly in the synthesis of ATP, but they were deficient to different extents in their supernumerary membrane subunits. In contrast, the enzymes from P. angusta and S. cerevisiae purified in the presence of n-decyl-β-maltose neopentyl glycol and the phospholipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, cardiolipin (diphosphatidylglycerol) and 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] had a complete complement of core subunits and also contained all of the known supernumerary membrane subunits, e, f, g, j, k and ATP8 (or Aap1), plus an additional new membrane component named subunit l, related in sequence to subunit k. The catalytic domain of the enzyme from P. angusta was more resistant to thermal denaturation than the enzyme from S. cerevisiae, but less stable than the catalytic domain of the bovine enzyme, but the stator and the integrity of the transmembrane proton pathway were most stable in the enzyme from P. angusta. The P. angusta enzyme provides a suitable source of enzyme for studying the structure of the membrane domain and properties associated with that sector of the enzyme complex.
Figures


Similar articles
-
Organization of Subunits in the Membrane Domain of the Bovine F-ATPase Revealed by Covalent Cross-linking.J Biol Chem. 2015 May 22;290(21):13308-20. doi: 10.1074/jbc.M115.645283. Epub 2015 Apr 7. J Biol Chem. 2015. PMID: 25851905 Free PMC article.
-
Dissociation and purification of the endogenous membrane-bound Vo complex from Pichia pastoris.Protein Expr Purif. 2017 Oct;138:76-80. doi: 10.1016/j.pep.2017.07.003. Epub 2017 Jul 12. Protein Expr Purif. 2017. PMID: 28709863
-
Two ATP synthases can be linked through subunits i in the inner mitochondrial membrane of Saccharomyces cerevisiae.Biochemistry. 2002 Aug 20;41(33):10390-6. doi: 10.1021/bi025923g. Biochemistry. 2002. PMID: 12173925
-
Structures and interactions of proteins involved in the coupling function of the protonmotive F(o)F(1)-ATP synthase.Curr Protein Pept Sci. 2002 Aug;3(4):451-60. doi: 10.2174/1389203023380558. Curr Protein Pept Sci. 2002. PMID: 12370007 Review.
-
Insights into ATP synthase assembly and function through the molecular genetic manipulation of subunits of the yeast mitochondrial enzyme complex.Biochim Biophys Acta. 2000 May 31;1458(2-3):428-42. doi: 10.1016/s0005-2728(00)00092-x. Biochim Biophys Acta. 2000. PMID: 10838056 Review.
Cited by
-
Structure of ATP synthase under strain during catalysis.Nat Commun. 2022 Apr 25;13(1):2232. doi: 10.1038/s41467-022-29893-2. Nat Commun. 2022. PMID: 35468906 Free PMC article.
-
Structure of the mitochondrial ATP synthase from Pichia angusta determined by electron cryo-microscopy.Proc Natl Acad Sci U S A. 2016 Nov 8;113(45):12709-12714. doi: 10.1073/pnas.1615902113. Epub 2016 Oct 24. Proc Natl Acad Sci U S A. 2016. PMID: 27791192 Free PMC article.
-
Structure of a catalytic dimer of the α- and β-subunits of the F-ATPase from Paracoccus denitrificans at 2.3 Å resolution.Acta Crystallogr F Struct Biol Commun. 2015 Oct;71(Pt 10):1309-17. doi: 10.1107/S2053230X15016076. Epub 2015 Sep 23. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 26457523 Free PMC article.
-
The Mitochondrial Permeability Transition Pore: Channel Formation by F-ATP Synthase, Integration in Signal Transduction, and Role in Pathophysiology.Physiol Rev. 2015 Oct;95(4):1111-55. doi: 10.1152/physrev.00001.2015. Physiol Rev. 2015. PMID: 26269524 Free PMC article. Review.
-
Purification, characterization and crystallization of the F-ATPase from Paracoccus denitrificans.Open Biol. 2015 Sep;5(9):150119. doi: 10.1098/rsob.150119. Open Biol. 2015. PMID: 26423580 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

