Similarities and differences of X and Y chromosome homologous genes, SRY and SOX3, in regulating the renin-angiotensin system promoters
- PMID: 25759379
- PMCID: PMC4421791
- DOI: 10.1152/physiolgenomics.00138.2014
Similarities and differences of X and Y chromosome homologous genes, SRY and SOX3, in regulating the renin-angiotensin system promoters
Abstract
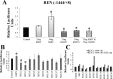
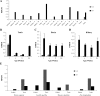
The renin-angiotensin system (RAS) is subject to sex-specific modulation by hormones and gene products. However, sex differences in the balance between the vasoconstrictor/proliferative ACE/ANG II/AT1 axis, and the vasodilator/antiproliferative ACE2/ANG-(1-7)/MAS axis are poorly known. Data in the rat have suggested the male-specific Y-chromosome gene Sry to contribute to balance between these two axes, but why the testis-determining gene has these functions remains unknown. A combination of in silico genetic/protein comparisons, functional luciferase assays for promoters of the human RAS, and RNA-Seq profiling in rat were used to address if regulation of Sry on the RAS is conserved in the homologous X-chromosome gene, Sox3. Both SRY and SOX3 upregulated the promoter of Angiotensinogen (AGT) and downregulated the promoters of ACE2, AT2, and MAS, likely through overlapping mechanisms. The regulation by both SRY and SOX3 on the MAS promoter indicates a cis regulation through multiple SOX binding sites. The Renin (REN) promoter is upregulated by SRY and downregulated by SOX3, likely through trans and cis mechanisms, respectively. Sry transcripts are found in all analyzed male rat tissues including the kidney, while Sox3 transcripts are found only in the brain and testis, suggesting that the primary tissue for renin production (kidney) can only be regulated by SRY and not SOX3. These results suggest that SRY regulation of the RAS is partially shared with its X-chromosome homolog SOX3, but SRY gained a sex-specific control in the kidney for the rate-limiting step of the RAS, potentially resulting in male-specific blood pressure regulation.
Keywords: Sox3; Sry; hypertension; renin angiotensin system; sex differences.
Copyright © 2015 the American Physiological Society.
Figures


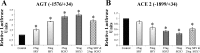

References
-
- Alatzoglou KS, Kelberman D, Dattani MT. The role of SOX proteins in normal pituitary development. J Endocrinol 200: 245–258, 2009. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 215: 403–410, 1990. - PubMed
-
- Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, Koutseva N, Zaghlul S, Graves T, Rock S, Kremitzki C, Fulton RS, Dugan S, Ding Y, Morton D, Khan Z, Lewis L, Buhay C, Wang Q, Watt J, Holder M, Lee S, Nazareth L, Rozen S, Muzny DM, Warren WC, Gibbs RA, Wilson RK, Page DC. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508: 494–499, 2014. - PMC - PubMed
-
- Berta P, Hawkins JB, Sinclair AH, Taylor A, Griffiths BL, Goodfellow PN, Fellous M. Genetic evidence equating SRY and the testis-determining factor. Nature 348: 448–450, 1990. - PubMed
-
- Bodiga VL, Bodiga S. Renin angiotensin system in cognitive function and dementia. Asian J Neurosci 2013: e102602, 2013.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

