Cleavage Specificity of Mycobacterium tuberculosis ClpP1P2 Protease and Identification of Novel Peptide Substrates and Boronate Inhibitors with Anti-bacterial Activity
- PMID: 25759383
- PMCID: PMC4409261
- DOI: 10.1074/jbc.M114.625640
Cleavage Specificity of Mycobacterium tuberculosis ClpP1P2 Protease and Identification of Novel Peptide Substrates and Boronate Inhibitors with Anti-bacterial Activity
Abstract
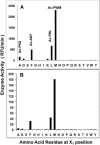
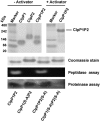
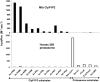
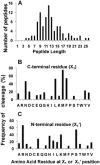
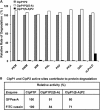
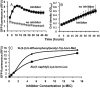
The ClpP1P2 protease complex is essential for viability in Mycobacteria tuberculosis and is an attractive drug target. Using a fluorogenic tripeptide library (Ac-X3X2X1-aminomethylcoumarin) and by determining specificity constants (kcat/Km), we show that ClpP1P2 prefers Met ≫ Leu > Phe > Ala in the X1 position, basic residues or Trp in the X2 position, and Pro ≫ Ala > Trp in the X3 position. We identified peptide substrates that are hydrolyzed up to 1000 times faster than the standard ClpP substrate. These positional preferences were consistent with cleavage sites in the protein GFPssrA by ClpXP1P2. Studies of ClpP1P2 with inactive ClpP1 or ClpP2 indicated that ClpP1 was responsible for nearly all the peptidase activity, whereas both ClpP1 and ClpP2 contributed to protein degradation. Substrate-based peptide boronates were synthesized that inhibit ClpP1P2 peptidase activity in the submicromolar range. Some of them inhibited the growth of Mtb cells in the low micromolar range indicating that cleavage specificity of Mtb ClpP1P2 can be used to design novel anti-bacterial agents.
Keywords: Drug Development; Enzyme Inhibitor; Enzyme Mechanism; Infectious Disease; Peptide Chemical Synthesis; Protein Degradation.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Raju R. M., Unnikrishnan M., Rubin D. H., Krishnamoorthy V., Kandror O., Akopian T. N., Goldberg A. L., Rubin E. J. (2012) Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS Pathog. 8, e1002511. - PMC - PubMed
-
- Sassetti C. M., Boyd D. H., Rubin E. J. (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 - PubMed
-
- Porankiewicz J., Wang J., Clarke A. K. (1999) New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32, 449–458 - PubMed
-
- Kang S. G., Dimitrova M. N., Ortega J., Ginsburg A., Maurizi M. R. (2005) Human mitochondrial ClpP is a stable heptamer that assembles into a tetradecamer in the presence of ClpX. J. Biol. Chem. 280, 35424–35432 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

