Origin, evolution, and virulence of porcine deltacoronaviruses in the United States
- PMID: 25759498
- PMCID: PMC4453528
- DOI: 10.1128/mBio.00064-15
Origin, evolution, and virulence of porcine deltacoronaviruses in the United States
Abstract
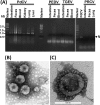
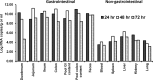
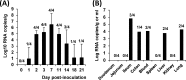
A novel porcine deltacoronavirus (PdCV) was first discovered in Ohio and Indiana in February 2014, rapidly spread to other states in the United States and Canada, and caused significant economic loss in the swine industry. The origin and virulence of this novel porcine coronavirus are not known. Here, we characterized U.S. PdCV isolates and determined their virulence in gnotobiotic and conventional piglets. Genome analyses revealed that U.S. PdCV isolates possess unique genetic characteristics and share a close relationship with Hong Kong and South Korean PdCV strains and coronaviruses (CoVs) of Asian leopard cats and Chinese ferret-badgers. The PdCV-positive intestinal content (Ohio CVM1) and the cell culture-adapted PdCV Michigan (MI) strain were orally inoculated into gnotobiotic and/or conventional piglets. Within 1 to 3 days postinfection, profuse watery diarrhea, vomiting, and dehydration were observed. Clinical signs were associated with epithelial necrosis in the gastric pits and small intestine, the latter resulting in severe villous atrophy. Mild interstitial pneumonia was identified in the lungs of PdCV-infected piglets. High levels of viral RNA (8 to 11 log RNA copies/g) were detected in intestinal tissues/luminal contents and feces of infected piglets, whereas moderate RNA levels (2 to 5 log RNA copies/g) were detected in blood, lung, liver, and kidney, indicating multisystemic dissemination of the virus. Polyclonal immune serum against PdCV but not immune serum against porcine epidemic diarrhea virus (PEDV) reacted with PdCV-infected small-intestinal epithelial cells, indicating that PdCV is antigenically distinct from PEDV. Collectively, we demonstrate for the first time that PdCV caused severe gastrointestinal diseases in swine.
Importance: Porcine coronaviruses (CoVs) are major viral infectious diseases of swine. Examples of porcine CoVs include porcine transmissible gastroenteritis coronavirus (TGEV), porcine epidemic diarrhea virus (PEDV), and porcine respiratory coronavirus (PRCV). In February 2014, another porcine CoV, porcine deltacoronavirus (PdCV), emerged in Ohio and Indiana and subsequently spread rapidly across the United States and Canada, causing significant economic losses. Here, we report the detailed genetic characterization, phylogeny, and virulence of emergent PdCV strains in the United States. We found that PdCV caused severe diarrhea, vomiting, and dehydration in gnotobiotic and conventional piglets, signs that were clinically indistinguishable from those caused by PEDV and TGEV. In addition to extensive intestinal lesions, PdCV caused significant lesions in the stomach and mild pulmonary lesions that have not been reported for TGEV and PEDV. The finding that PdCV is a significant enteric disease of swine highlights the need to develop effective measures to control this disease.
Copyright © 2015 Ma et al.
Figures





Similar articles
-
Deletion of a 197-Amino-Acid Region in the N-Terminal Domain of Spike Protein Attenuates Porcine Epidemic Diarrhea Virus in Piglets.J Virol. 2017 Jun 26;91(14):e00227-17. doi: 10.1128/JVI.00227-17. Print 2017 Jul 15. J Virol. 2017. PMID: 28490591 Free PMC article.
-
Emerging and re-emerging coronaviruses in pigs.Curr Opin Virol. 2019 Feb;34:39-49. doi: 10.1016/j.coviro.2018.12.001. Epub 2019 Jan 14. Curr Opin Virol. 2019. PMID: 30654269 Free PMC article. Review.
-
A Highly Pathogenic Strain of Porcine Deltacoronavirus Caused Watery Diarrhea in Newborn Piglets.Virol Sin. 2018 Apr;33(2):131-141. doi: 10.1007/s12250-018-0003-8. Epub 2018 Mar 22. Virol Sin. 2018. PMID: 29569144 Free PMC article.
-
Porcine deltacoronavirus infection: Etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis.Virus Res. 2016 Dec 2;226:50-59. doi: 10.1016/j.virusres.2016.04.009. Epub 2016 Apr 13. Virus Res. 2016. PMID: 27086031 Free PMC article. Review.
-
Infection, genetic and virulence characteristics of porcine epidemic diarrhea virus in northwest China.Infect Genet Evol. 2018 Aug;62:34-39. doi: 10.1016/j.meegid.2018.04.001. Epub 2018 Apr 4. Infect Genet Evol. 2018. PMID: 29625238
Cited by
-
Isolation and Tissue Culture Adaptation of Porcine Deltacoronavirus: A Case Study.Methods Mol Biol. 2020;2203:77-88. doi: 10.1007/978-1-0716-0900-2_6. Methods Mol Biol. 2020. PMID: 32833205
-
Genome-Wide CRISPR/Cas9 Screen Reveals a Role for SLC35A1 in the Adsorption of Porcine Deltacoronavirus.J Virol. 2022 Dec 21;96(24):e0162622. doi: 10.1128/jvi.01626-22. Epub 2022 Dec 1. J Virol. 2022. PMID: 36453883 Free PMC article.
-
Cross-species transmission, evolution and zoonotic potential of coronaviruses.Front Cell Infect Microbiol. 2023 Jan 6;12:1081370. doi: 10.3389/fcimb.2022.1081370. eCollection 2022. Front Cell Infect Microbiol. 2023. PMID: 36683695 Free PMC article. Review.
-
Aminopeptidase N-null neonatal piglets are protected from transmissible gastroenteritis virus but not porcine epidemic diarrhea virus.Sci Rep. 2019 Sep 12;9(1):13186. doi: 10.1038/s41598-019-49838-y. Sci Rep. 2019. PMID: 31515498 Free PMC article.
-
Genomic characterization and pathogenicity of porcine deltacoronavirus strain CHN-HG-2017 from China.Arch Virol. 2019 Feb;164(2):413-425. doi: 10.1007/s00705-018-4081-6. Epub 2018 Oct 30. Arch Virol. 2019. PMID: 30377826 Free PMC article.
References
-
- Doyle LP, Hutchings LM. 1946. A transmissible gastroenteritis in pigs. J Am Vet Med Assoc 108:257–259. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

