The Open Physiology workflow: modeling processes over physiology circuitboards of interoperable tissue units
- PMID: 25759670
- PMCID: PMC4338662
- DOI: 10.3389/fphys.2015.00024
The Open Physiology workflow: modeling processes over physiology circuitboards of interoperable tissue units
Abstract
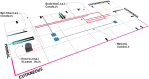
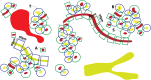
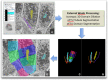
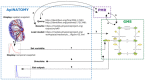
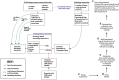
A key challenge for the physiology modeling community is to enable the searching, objective comparison and, ultimately, re-use of models and associated data that are interoperable in terms of their physiological meaning. In this work, we outline the development of a workflow to modularize the simulation of tissue-level processes in physiology. In particular, we show how, via this approach, we can systematically extract, parcellate and annotate tissue histology data to represent component units of tissue function. These functional units are semantically interoperable, in terms of their physiological meaning. In particular, they are interoperable with respect to [i] each other and with respect to [ii] a circuitboard representation of long-range advective routes of fluid flow over which to model long-range molecular exchange between these units. We exemplify this approach through the combination of models for physiology-based pharmacokinetics and pharmacodynamics to quantitatively depict biological mechanisms across multiple scales. Links to the data, models and software components that constitute this workflow are found at http://open-physiology.org/.
Keywords: ApiNATOMY; fluid flow modeling; functional tissue units; histology; physiology circuit-boarding; physiology-based pharmacokinetics; visual knowledge management.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

