Effects on murine behavior and lifespan of selectively decreasing expression of mutant huntingtin allele by supt4h knockdown
- PMID: 25760041
- PMCID: PMC4356588
- DOI: 10.1371/journal.pgen.1005043
Effects on murine behavior and lifespan of selectively decreasing expression of mutant huntingtin allele by supt4h knockdown
Abstract
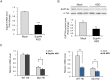
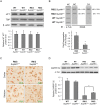
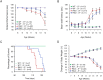
Production of protein containing lengthy stretches of polyglutamine encoded by multiple repeats of the trinucleotide CAG is a hallmark of Huntington's disease (HD) and of a variety of other inherited degenerative neurological and neuromuscular disorders. Earlier work has shown that interference with production of the transcription elongation protein SUPT4H results in decreased cellular capacity to transcribe mutant huntingtin gene (Htt) alleles containing long CAG expansions, but has little effect on expression of genes containing short CAG stretches. zQ175 and R6/2 are genetically engineered mouse strains whose genomes contain human HTT alleles that include greatly expanded CAG repeats and which are used as animal models for HD. Here we show that reduction of SUPT4H expression in brains of zQ175 mice by intracerebroventricular bolus injection of antisense 2'-O-methoxyethyl oligonucleotides (ASOs) directed against Supt4h, or in R6/2 mice by deletion of one copy of the Supt4h gene, results in a decrease in mRNA and protein encoded specifically by mutant Htt alleles. We further show that reduction of SUPT4H in mouse brains is associated with decreased HTT protein aggregation, and in R6/2 mice, also with prolonged lifespan and delay of the motor impairment that normally develops in these animals. Our findings support the view that targeting of SUPT4H function may be useful as a therapeutic countermeasure against HD.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Ashley CT Jr., Warren ST (1995) Trinucleotide repeat expansion and human disease. Annu Rev Genet 29: 703–728. - PubMed
-
- Orr HT, Zoghbi HY (2007) Trinucleotide repeat disorders. Annu Rev Neurosci 30: 575–621. - PubMed
-
- Group. HsDCR (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell 72: 971–983. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

