2.8 Å resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy
- PMID: 25760083
- PMCID: PMC4391500
- DOI: 10.7554/eLife.06380
2.8 Å resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy
Abstract
Recent developments in detector hardware and image-processing software have revolutionized single particle cryo-electron microscopy (cryoEM) and led to a wave of near-atomic resolution (typically ∼3.3 Å) reconstructions. Reaching resolutions higher than 3 Å is a prerequisite for structure-based drug design and for cryoEM to become widely interesting to pharmaceutical industries. We report here the structure of the 700 kDa Thermoplasma acidophilum 20S proteasome (T20S), determined at 2.8 Å resolution by single-particle cryoEM. The quality of the reconstruction enables identifying the rotameric conformation adopted by some amino-acid side chains (rotamers) and resolving ordered water molecules, in agreement with the expectations for crystal structures at similar resolutions. The results described in this manuscript demonstrate that single particle cryoEM is capable of competing with X-ray crystallography for determination of protein structures of suitable quality for rational drug design.
Keywords: T. acidophilum; biophysics; direct detectors; electron microscopy; image processing; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
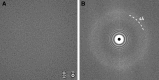

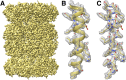
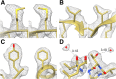
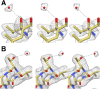

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

