Expression pattern and subcellular localization of the ovate protein family in rice
- PMID: 25760462
- PMCID: PMC4356581
- DOI: 10.1371/journal.pone.0118966
Expression pattern and subcellular localization of the ovate protein family in rice
Abstract
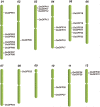
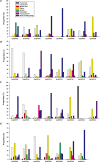
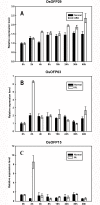
The Arabidopsis ovate family proteins (AtOFPs) have been shown to function as transcriptional repressors and regulate multiple aspects of plant growth and development. There are 31 genes that encode the full-length OVATE-domain containing proteins in the rice genome. In this study, the gene structure analysis revealed that OsOFPs are intron poor. Phylogenetic analysis suggested that OVATE proteins from rice, Arabidopsis and tomato can be divided into 4 groups (I-IV). Real-time quantitative polymerase chain reaction (RT-qPCR) analysis identified OsOFPs with different tissue-specific expression patterns at all stages of development in the rice plant. Interestingly, nearly half of the total number of OsOFP family was more highly expressed during the seed developmental stage. In addition, seed developmental cis-elements were found in the promoter region of the OsOFPs. Subcellular localization analysis revealed that YFP-OsOFP fusion proteins predominantly localized in the nucleus. Our results suggest that OsOFPs may act as regulatory proteins and play pivotal roles in the growth and development of rice.
Conflict of interest statement
Figures



Similar articles
-
Overview of OVATE FAMILY PROTEINS, A Novel Class of Plant-Specific Growth Regulators.Front Plant Sci. 2016 Mar 31;7:417. doi: 10.3389/fpls.2016.00417. eCollection 2016. Front Plant Sci. 2016. PMID: 27065353 Free PMC article. Review.
-
Phylogenetic analyses provide the first insights into the evolution of OVATE family proteins in land plants.Ann Bot. 2014 Jun;113(7):1219-33. doi: 10.1093/aob/mcu061. Epub 2014 May 8. Ann Bot. 2014. PMID: 24812252 Free PMC article.
-
Plant omics: genome-wide analysis of ABA repressor1 (ABR1) related genes in rice during abiotic stress and development.OMICS. 2013 Aug;17(8):439-50. doi: 10.1089/omi.2012.0074. OMICS. 2013. PMID: 23895290
-
Rice OVATE family protein 6 regulates leaf angle by modulating secondary cell wall biosynthesis.Plant Mol Biol. 2020 Oct;104(3):249-261. doi: 10.1007/s11103-020-01039-2. Epub 2020 Jul 26. Plant Mol Biol. 2020. PMID: 32715397
-
The shape of things to come: ovate family proteins regulate plant organ shape.Curr Opin Plant Biol. 2020 Feb;53:98-105. doi: 10.1016/j.pbi.2019.10.005. Epub 2019 Dec 11. Curr Opin Plant Biol. 2020. PMID: 31837627 Review.
Cited by
-
Identification, Expression, and Interaction Analysis of Ovate Family Proteins in Populus trichocarpa Reveals a Role of PtOFP1 Regulating Drought Stress Response.Front Plant Sci. 2021 Apr 20;12:650109. doi: 10.3389/fpls.2021.650109. eCollection 2021. Front Plant Sci. 2021. PMID: 33959141 Free PMC article.
-
Overview of OVATE FAMILY PROTEINS, A Novel Class of Plant-Specific Growth Regulators.Front Plant Sci. 2016 Mar 31;7:417. doi: 10.3389/fpls.2016.00417. eCollection 2016. Front Plant Sci. 2016. PMID: 27065353 Free PMC article. Review.
-
Quantitative phosphoproteomic analysis of early seed development in rice (Oryza sativa L.).Plant Mol Biol. 2016 Feb;90(3):249-65. doi: 10.1007/s11103-015-0410-2. Epub 2015 Nov 28. Plant Mol Biol. 2016. PMID: 26613898
-
Genome-wide identification and characterization of the OFP gene family in Chinese cabbage (Brassica rapa L. ssp. pekinensis).PeerJ. 2021 Mar 5;9:e10934. doi: 10.7717/peerj.10934. eCollection 2021. PeerJ. 2021. PMID: 33717690 Free PMC article.
-
Identification and Comprehensive Analysis of OFP Genes for Fruit Shape Influence in Mango.Genes (Basel). 2024 Jun 21;15(7):823. doi: 10.3390/genes15070823. Genes (Basel). 2024. PMID: 39062602 Free PMC article.
References
-
- Scott MP. Development: the natural history of genes. Cell. 2000;100: 27–40. - PubMed
-
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290: 2105–2110. - PubMed
-
- Guo A, He K, Liu D, Bai S, Gu X, et al. DATF: a database of Arabidopsis transcription factors. Bioinformatics. 2005;21: 2568–2569. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

