Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase
- PMID: 25760614
- PMCID: PMC4356371
- DOI: 10.1107/S1399004715000152
Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase
Abstract
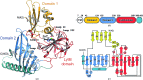
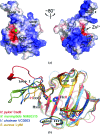
Helicobacter pylori is associated with various gastrointestinal diseases such as gastritis, ulcers and gastric cancer. Its colonization of the human gastric mucosa requires high motility, which depends on its helical cell shape. Seven cell shape-determining genes (csd1, csd2, csd3/hdpA, ccmA, csd4, csd5 and csd6) have been identified in H. pylori. Their proteins play key roles in determining the cell shape through modifications of the cell-wall peptidoglycan by the alteration of cross-linking or by the trimming of peptidoglycan muropeptides. Among them, Csd3 (also known as HdpA) is a bifunctional enzyme. Its D,D-endopeptidase activity cleaves the D-Ala(4)-mDAP(3) peptide bond between cross-linked muramyl tetrapeptides and pentapeptides. It is also a D,D-carboxypeptidase that cleaves off the terminal D-Ala(5) from the muramyl pentapeptide. Here, the crystal structure of this protein has been determined, revealing the organization of its three domains in a latent and inactive state. The N-terminal domain 1 and the core of domain 2 share the same fold despite a very low level of sequence identity, and their surface-charge distributions are different. The C-terminal LytM domain contains the catalytic site with a Zn(2+) ion, like the similar domains of other M23 metallopeptidases. Domain 1 occludes the active site of the LytM domain. The core of domain 2 is held against the LytM domain by the C-terminal tail region that protrudes from the LytM domain.
Keywords: HP0506; Helicobacter pylori; LytM; M23B family metallopeptidase; cell-shape determinant; csd3; d,d-carboxypeptidase; d,d-endopeptidase; peptidoglycan hydrolase.
Figures




References
-
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221. - PubMed
-
- Berg, H. C. & Turner, L. (1979). Nature (London), 278, 349–351. - PubMed
-
- Bonis, M., Ecobichon, C., Guadagnini, S., Prévost, M.-C. & Boneca, I. G. (2010). Mol. Microbiol. 78, 809–819. - PubMed
-
- Brünger, A. T. (1992). Nature (London), 355, 472–475. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

