HIV broadly neutralizing antibody targets
- PMID: 25760932
- PMCID: PMC4437463
- DOI: 10.1097/COH.0000000000000153
HIV broadly neutralizing antibody targets
Abstract
Purpose of review: To provide an update on neutralizing antibody targets in the context of the recent HIV-1 envelope trimer structure, describe new antibody isolation technologies, and discuss the implications of these data for HIV-1 prevention and therapy.
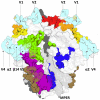
Recent findings: Recent advances in B-cell technologies have dramatically expanded the number of antibodies isolated from HIV-infected donors with broadly neutralizing plasma activity. These, together with the first high-resolution crystal and cryo-electron microscopy (cryo-EM) structures of a cleaved, prefusion HIV-1 trimer, have defined new regions susceptible to neutralization. This year, three epitopes in the gp120-gp41 interface were structurally characterized, highlighting the importance of prefusion gp41 as a target. Similar to many other broadly neutralizing antibody epitopes, these new antibodies define a target that is also highly glycan dependent. Collectively, the epitopes for broadly neutralizing antibodies now reveal a continuum of vulnerability spanning the length of the HIV-1 envelope trimer.
Summary: Progress in the last year has provided support for the use of rationally stabilized whole HIV-1 trimers as immunogens for eliciting antibodies to multiple epitopes. Furthermore, the increasing number of broad and potent antibodies with the potential for synergistic/complementary combinations opens up new avenues for preventing and treating HIV-1 infection.
Figures


References
-
-
Pancera M, Zhou T, Druz A, et al. Structure and immune recognition of trimeric prefusion HIV-1 Env. Nature. 2014;514:455–461. Using 35O22 and PGT122, the authors of this article crystallized the Env trimer in near entirety, building on work by Julien et al., 2013 [4], by resolving gp41. These data revealed the interactions that stabilize the prefusion trimer, and showed how HIV-1 Env proteins share remarkable mechanistic similarity with respiratory syncytial virus, Ebola, and influenza. The complex with 35O22 revealed binding to N625 that was incompatible with a glycan covalently linked to this side chain.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

