The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family
- PMID: 25761566
- PMCID: PMC4356964
- DOI: 10.1038/srep09040
The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family
Abstract
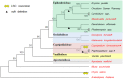
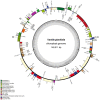
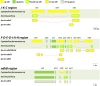
The NAD(P)H dehydrogenase complex is encoded by 11 ndh genes in plant chloroplast (cp) genomes. However, ndh genes are truncated or deleted in some autotrophic Epidendroideae orchid cp genomes. To determine the evolutionary timing of the gene deletions and the genomic locations of the various ndh genes in orchids, the cp genomes of Vanilla planifolia, Paphiopedilum armeniacum, Paphiopedilum niveum, Cypripedium formosanum, Habenaria longidenticulata, Goodyera fumata and Masdevallia picturata were sequenced; these genomes represent Vanilloideae, Cypripedioideae, Orchidoideae and Epidendroideae subfamilies. Four orchid cp genome sequences were found to contain a complete set of ndh genes. In other genomes, ndh deletions did not correlate to known taxonomic or evolutionary relationships and deletions occurred independently after the orchid family split into different subfamilies. In orchids lacking cp encoded ndh genes, non cp localized ndh sequences were identified. In Erycina pusilla, at least 10 truncated ndh gene fragments were found transferred to the mitochondrial (mt) genome. The phenomenon of orchid ndh transfer to the mt genome existed in ndh-deleted orchids and also in ndh containing species.
Figures



References
-
- Archibald J. M. Origin of eukaryotic cells: 40 years on. Symbiosis 54, 69–86 (2011).
-
- Fuentes I., Karcher D. & Bock R. Experimental reconstruction of the functional transfer of intron-containing plastid genes to the nucleus. Curr. Biol. 22, 763–771 (2012). - PubMed
-
- Zurawski G. & Clegg M. T. Evolution of higher-plant chloroplast DNA-encoded genes: implications for structure-function and phylogenetic studies. Ann. Rev. Plant Physiol. 38, 391–418 (1987).
-
- Matsubayashi T. et al. Six chloroplast genes (ndhA-F) homologous to human mitochondrial genes encoding components of the respiratory chain NADH dehydrogenase are actively expressed: determination of the splice sites in ndhA and ndhB pre-mRNAs. Mol. Gen. Genet. 210, 385–393 (1987). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

