Effect of minichromosome maintenance protein 2 deficiency on the locations of DNA replication origins
- PMID: 25762552
- PMCID: PMC4381527
- DOI: 10.1101/gr.176099.114
Effect of minichromosome maintenance protein 2 deficiency on the locations of DNA replication origins
Abstract
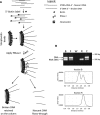
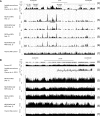
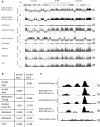
Minichromosome maintenance (MCM) proteins are loaded onto chromatin during G1-phase and define potential locations of DNA replication initiation. MCM protein deficiency results in genome instability and high rates of cancer in mouse models. Here we develop a method of nascent strand capture and release and show that MCM2 deficiency reduces DNA replication initiation in gene-rich regions of the genome. DNA structural properties are shown to correlate with sequence motifs associated with replication origins and with locations that are preferentially affected by MCM2 deficiency. Reduced nascent strand density correlates with sites of recurrent focal CNVs in tumors arising in MCM2-deficient mice, consistent with a direct relationship between sites of reduced DNA replication initiation and genetic damage. Between 10% and 90% of human tumors, depending on type, carry heterozygous loss or mutation of one or more MCM2-7 genes, which is expected to compromise DNA replication origin licensing and result in elevated rates of genome damage at a subset of gene-rich locations.
© 2015 Kunnev et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin JM, Lemaitre JM. 2012. Unraveling cell type–specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol 19: 837–844. - PubMed
-
- Bielinsky AK, Gerbi SA. 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279: 95–98. - PubMed
Publication types
MeSH terms
Substances
Associated data
- BioProject/PRJNA258088
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
