Salmonella serotype determination utilizing high-throughput genome sequencing data
- PMID: 25762776
- PMCID: PMC4400759
- DOI: 10.1128/JCM.00323-15
Salmonella serotype determination utilizing high-throughput genome sequencing data
Abstract
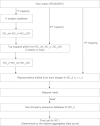
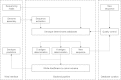
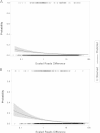
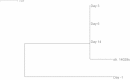
Serotyping forms the basis of national and international surveillance networks for Salmonella, one of the most prevalent foodborne pathogens worldwide (1-3). Public health microbiology is currently being transformed by whole-genome sequencing (WGS), which opens the door to serotype determination using WGS data. SeqSero (www.denglab.info/SeqSero) is a novel Web-based tool for determining Salmonella serotypes using high-throughput genome sequencing data. SeqSero is based on curated databases of Salmonella serotype determinants (rfb gene cluster, fliC and fljB alleles) and is predicted to determine serotype rapidly and accurately for nearly the full spectrum of Salmonella serotypes (more than 2,300 serotypes), from both raw sequencing reads and genome assemblies. The performance of SeqSero was evaluated by testing (i) raw reads from genomes of 308 Salmonella isolates of known serotype; (ii) raw reads from genomes of 3,306 Salmonella isolates sequenced and made publicly available by GenomeTrakr, a U.S. national monitoring network operated by the Food and Drug Administration; and (iii) 354 other publicly available draft or complete Salmonella genomes. We also demonstrated Salmonella serotype determination from raw sequencing reads of fecal metagenomes from mice orally infected with this pathogen. SeqSero can help to maintain the well-established utility of Salmonella serotyping when integrated into a platform of WGS-based pathogen subtyping and characterization.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
References
-
- Grimont P, Weill F. 2007. Antigenic formulae of the Salmonella serovars, 9th ed WHO Collaborating Centre for Reference and Research on Salmonella. WHO, Geneva, Switzerland.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

