Transcriptional Adaptation of Drug-tolerant Mycobacterium tuberculosis During Treatment of Human Tuberculosis
- PMID: 25762787
- PMCID: PMC4548467
- DOI: 10.1093/infdis/jiv149
Transcriptional Adaptation of Drug-tolerant Mycobacterium tuberculosis During Treatment of Human Tuberculosis
Abstract
Background: Treatment initiation rapidly kills most drug-susceptible Mycobacterium tuberculosis, but a bacterial subpopulation tolerates prolonged drug exposure. We evaluated drug-tolerant bacilli in human sputum by comparing messenger RNA (mRNA) expression of drug-tolerant bacilli that survive the early bactericidal phase with treatment-naive bacilli.
Methods: M. tuberculosis gene expression was quantified via reverse-transcription polymerase chain reaction in serial sputa from 17 Ugandans treated for drug-susceptible pulmonary tuberculosis.
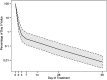
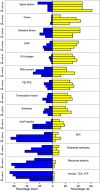
Results: Within 4 days, bacterial mRNA abundance declined >98%, indicating rapid killing. Thereafter, the rate of decline slowed >94%, indicating drug tolerance. After 14 days, 16S ribosomal RNA transcripts/genome declined 96%, indicating slow growth. Drug-tolerant bacilli displayed marked downregulation of genes associated with growth, metabolism, and lipid synthesis and upregulation in stress responses and key regulatory categories-including stress-associated sigma factors, transcription factors, and toxin-antitoxin genes. Drug efflux pumps were upregulated. The isoniazid stress signature was induced by initial drug exposure, then disappeared after 4 days.
Conclusions: Transcriptional patterns suggest that drug-tolerant bacilli in sputum are in a slow-growing, metabolically and synthetically downregulated state. Absence of the isoniazid stress signature in drug-tolerant bacilli indicates that physiological state influences drug responsiveness in vivo. These results identify novel drug targets that should aid in development of novel shorter tuberculosis treatment regimens.
Keywords: Mycobacterium tuberculosis/genetics; Mycobacterium tuberculosis/physiology; gene expression profiling; pulmonary/epidemiology; sputum/microbiology; tuberculosis.
© The Author 2015. Published by Oxford University Press on behalf of the Infectious Diseases Society of America. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- World Health Organization. Global tuberculosis control: WHO report 2014. 2014.
-
- Mitchison DA, Selkon JB. The bactericidal activities of antituberculosis drugs. Am Rev Respir Dis 1956; 74:109–16. - PubMed
-
- Jindani A, Doré CJ, Mitchison DA. Bactericidal and sterilizing activities of antituberculosis drugs during the first 14 Days. Am J Resp Crit Care 2003; 167:1348–54. - PubMed
-
- Balaban NQ, Gerdes K, Lewis K, McKinney JD. A problem of persistence: still more questions than answers? Nat Rev Micro 2013; 11:587–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI061505/AI/NIAID NIH HHS/United States
- K24HL087713/HL/NHLBI NIH HHS/United States
- T15 LM009451/LM/NLM NIH HHS/United States
- 2T15LM009451-06/LM/NLM NIH HHS/United States
- R01 AI104589/AI/NIAID NIH HHS/United States
- K23 AI080147/AI/NIAID NIH HHS/United States
- K23 HL094141/HL/NHLBI NIH HHS/United States
- UL1 RR025780/RR/NCRR NIH HHS/United States
- 2R01 AI061505/AI/NIAID NIH HHS/United States
- R21 AI101714/AI/NIAID NIH HHS/United States
- K23AI080147/AI/NIAID NIH HHS/United States
- K24 HL087713/HL/NHLBI NIH HHS/United States
- K23HL094141/HL/NHLBI NIH HHS/United States
- R01 HL090335/HL/NHLBI NIH HHS/United States
- R01HL090335/HL/NHLBI NIH HHS/United States
- UL1 TR001082/TR/NCATS NIH HHS/United States
- R21AI101714/AI/NIAID NIH HHS/United States
- 5R01AI104589/AI/NIAID NIH HHS/United States
- IK2 CX000914/CX/CSRD VA/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

