Contributions of in vitro transcription to the understanding of human RNA polymerase III transcription
- PMID: 25764111
- PMCID: PMC4214236
- DOI: 10.4161/trns.27526
Contributions of in vitro transcription to the understanding of human RNA polymerase III transcription
Abstract
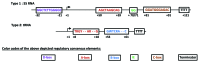
Human RNA polymerase III transcribes small untranslated RNAs that contribute to the regulation of essential cellular processes, including transcription, RNA processing and translation. Analysis of this transcription system by in vitro transcription techniques has largely contributed to the discovery of its transcription factors and to the understanding of the regulation of human RNA polymerase III transcription. Here we review some of the key steps that led to the identification of transcription factors and to the definition of minimal promoter sequences for human RNA polymerase III transcription.
Keywords: Human RNA polymerase III; PTF; SNAPC; TFIIIA; TFIIIB; TFIIIC; activated transcription; basal transcription; enhancer; in vitro; promoter.
Figures


References
-
- Weil PA, Segall J, Harris B, Ng SY, Roeder RG. Faithful transcription of eukaryotic genes by RNA polymerase III in systems reconstituted with purified DNA templates. J Biol Chem. 1979;254:6163–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
