Transcription elongation. Heterogeneous tracking of RNA polymerase and its biological implications
- PMID: 25764114
- PMCID: PMC4214235
- DOI: 10.4161/trns.28285
Transcription elongation. Heterogeneous tracking of RNA polymerase and its biological implications
Abstract
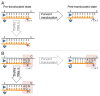
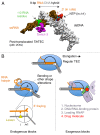
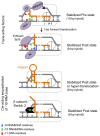
Regulation of transcription elongation via pausing of RNA polymerase has multiple physiological roles. The pausing mechanism depends on the sequence heterogeneity of the DNA being transcribed, as well as on certain interactions of polymerase with specific DNA sequences. In order to describe the mechanism of regulation, we introduce the concept of heterogeneity into the previously proposed alternative models of elongation, power stroke and Brownian ratchet. We also discuss molecular origins and physiological significances of the heterogeneity.
Keywords: Brownian ratchet; RNA polymerase; power stroke; sequence heterogeneity of DNA; transcription elongation factors; transcription pausing; translocation.
Figures
Similar articles
-
Pause & go: from the discovery of RNA polymerase pausing to its functional implications.Curr Opin Cell Biol. 2017 Jun;46:72-80. doi: 10.1016/j.ceb.2017.03.002. Epub 2017 Mar 28. Curr Opin Cell Biol. 2017. PMID: 28363125 Free PMC article. Review.
-
Translocation by T7 RNA polymerase: a sensitively poised Brownian ratchet.J Mol Biol. 2006 Apr 21;358(1):241-54. doi: 10.1016/j.jmb.2006.02.001. Epub 2006 Feb 14. J Mol Biol. 2006. PMID: 16516229
-
A dynamic model for transcription elongation and sequence-dependent short pauses by RNA polymerase.Biosystems. 2008 Sep;93(3):199-210. doi: 10.1016/j.biosystems.2008.04.013. Epub 2008 May 4. Biosystems. 2008. PMID: 18539382
-
Transcription pausing: biological significance of thermal fluctuations biased by repetitive genomic sequences.Transcription. 2018;9(3):196-203. doi: 10.1080/21541264.2017.1393492. Epub 2017 Dec 1. Transcription. 2018. PMID: 29105534 Free PMC article.
-
[RNA polymerase as a molecular machine].Mol Biol (Mosk). 2002 Mar-Apr;36(2):208-15. Mol Biol (Mosk). 2002. PMID: 11969082 Review. Russian.
Cited by
-
Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo.Genome Biol. 2015 May 15;16(1):98. doi: 10.1186/s13059-015-0666-5. Genome Biol. 2015. PMID: 25976475 Free PMC article.
-
RNA polymerase II transcriptional fidelity control and its functional interplay with DNA modifications.Crit Rev Biochem Mol Biol. 2015;50(6):503-19. doi: 10.3109/10409238.2015.1087960. Epub 2015 Sep 22. Crit Rev Biochem Mol Biol. 2015. PMID: 26392149 Free PMC article. Review.
-
Pause & go: from the discovery of RNA polymerase pausing to its functional implications.Curr Opin Cell Biol. 2017 Jun;46:72-80. doi: 10.1016/j.ceb.2017.03.002. Epub 2017 Mar 28. Curr Opin Cell Biol. 2017. PMID: 28363125 Free PMC article. Review.
-
Conserved DNA sequence features underlie pervasive RNA polymerase pausing.Nucleic Acids Res. 2021 May 7;49(8):4402-4420. doi: 10.1093/nar/gkab208. Nucleic Acids Res. 2021. PMID: 33788942 Free PMC article.
-
RNAs as Regulators of Cellular Matchmaking.Front Mol Biosci. 2021 Apr 9;8:634146. doi: 10.3389/fmolb.2021.634146. eCollection 2021. Front Mol Biosci. 2021. PMID: 33898516 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
