SARS hCoV papain-like protease is a unique Lys48 linkage-specific di-distributive deubiquitinating enzyme
- PMID: 25764917
- PMCID: PMC4447217
- DOI: 10.1042/BJ20141170
SARS hCoV papain-like protease is a unique Lys48 linkage-specific di-distributive deubiquitinating enzyme
Abstract
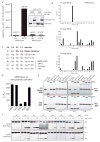
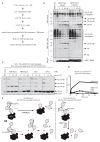
Ubiquitin (Ub) and the Ub-like (Ubl) modifier interferon-stimulated gene 15 (ISG15) participate in the host defence of viral infections. Viruses, including the severe acute respiratory syndrome human coronavirus (SARS hCoV), have co-opted Ub-ISG15 conjugation pathways for their own advantage or have evolved effector proteins to counter pro-inflammatory properties of Ub-ISG15-conjugated host proteins. In the present study, we compare substrate specificities of the papain-like protease (PLpro) from the recently emerged Middle East respiratory syndrome (MERS) hCoV to the related protease from SARS, SARS PLpro. Through biochemical assays, we show that, similar to SARS PLpro, MERS PLpro is both a deubiquitinating (DUB) and a deISGylating enzyme. Further analysis of the intrinsic DUB activity of these viral proteases revealed unique differences between the recognition and cleavage specificities of polyUb chains. First, MERS PLpro shows broad linkage specificity for the cleavage of polyUb chains, whereas SARS PLpro prefers to cleave Lys48-linked polyUb chains. Secondly, MERS PLpro cleaves polyUb chains in a 'mono-distributive' manner (one Ub at a time) and SARS PLpro prefers to cleave Lys48-linked polyUb chains by sensing a di-Ub moiety as a minimal recognition element using a 'di-distributive' cleavage mechanism. The di-distributive cleavage mechanism for SARS PLpro appears to be uncommon among USP (Ub-specific protease)-family DUBs, as related USP family members from humans do not display such a mechanism. We propose that these intrinsic enzymatic differences between SARS and MERS PLpro will help to identify pro-inflammatory substrates of these viral DUBs and can guide in the design of therapeutics to combat infection by coronaviruses.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Herfst S, Fouchier R. Epidemiological and genetic investigations of human-to-human transmission of zoonotic influenza viruses. Euro Surveill 2014. 2014;19(25) pii=20840. - PubMed
-
- Baez-Santos YM, Barraza SJ, Wilson MW, Agius MP, Mielech AM, Davis NM, Baker SC, Larsen SD, Mesecar AD. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. Journal of Medicinal Chemistry. 2014;57:2393–2412. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

