Vaccinia virus protein A3 is required for the production of normal immature virions and for the encapsidation of the nucleocapsid protein L4
- PMID: 25765002
- PMCID: PMC4437912
- DOI: 10.1016/j.virol.2015.02.020
Vaccinia virus protein A3 is required for the production of normal immature virions and for the encapsidation of the nucleocapsid protein L4
Abstract
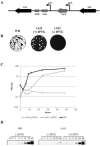
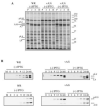
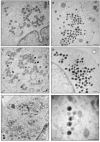
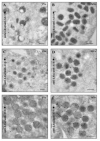
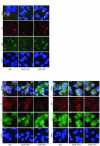
Maturation of the vaccinia virion is an intricate process that results in the organization of the viroplasm contained in immature virions into the lateral bodies, core wall and nucleocapsid observed in the mature particles. It is unclear how this organization takes place and studies with mutants are indispensable in understanding this process. By characterizing an inducible mutant in the A3L gene, we revealed that A3, an inner core wall protein, is important for formation of normal immature viruses and also for the correct localization of L4, a nucleocapsid protein. L4 did not accumulate in the viral factories in the absence of A3 and was not encapsidated in the particles that do not contain A3. These data strengthen our previously suggested hypothesis that A3 and L4 interact and that this interaction is critical for proper formation of the core wall and nucleocapsid.
Keywords: A3; Core wall; Nucleocapsid; Vaccinia virus.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
The vaccinia virus E6 protein influences virion protein localization during virus assembly.Virology. 2015 Aug;482:147-56. doi: 10.1016/j.virol.2015.02.056. Epub 2015 Apr 10. Virology. 2015. PMID: 25863879 Free PMC article.
-
Vaccinia virus mutations in the L4R gene encoding a virion structural protein produce abnormal mature particles lacking a nucleocapsid.J Virol. 2014 Dec;88(24):14017-29. doi: 10.1128/JVI.02126-14. Epub 2014 Sep 24. J Virol. 2014. PMID: 25253347 Free PMC article.
-
Vaccinia virus A6 is essential for virion membrane biogenesis and localization of virion membrane proteins to sites of virion assembly.J Virol. 2012 May;86(10):5603-13. doi: 10.1128/JVI.00330-12. Epub 2012 Mar 7. J Virol. 2012. PMID: 22398288 Free PMC article.
-
From crescent to mature virion: vaccinia virus assembly and maturation.Viruses. 2014 Oct 7;6(10):3787-808. doi: 10.3390/v6103787. Viruses. 2014. PMID: 25296112 Free PMC article. Review.
-
In a nutshell: structure and assembly of the vaccinia virion.Adv Virus Res. 2006;66:31-124. doi: 10.1016/S0065-3527(06)66002-8. Adv Virus Res. 2006. PMID: 16877059 Review.
Cited by
-
Transcriptome Changes in Glioma Cells upon Infection with the Oncolytic Virus VV-GMCSF-Lact.Cells. 2023 Nov 12;12(22):2616. doi: 10.3390/cells12222616. Cells. 2023. PMID: 37998351 Free PMC article.
-
mTOR Dysregulation by Vaccinia Virus F17 Controls Multiple Processes with Varying Roles in Infection.J Virol. 2019 Jul 17;93(15):e00784-19. doi: 10.1128/JVI.00784-19. Print 2019 Aug 1. J Virol. 2019. PMID: 31118254 Free PMC article.
-
Real-Time Capture and Visualization of Individual Viruses in Complex Media.ACS Nano. 2016 Feb 23;10(2):2827-33. doi: 10.1021/acsnano.5b07948. Epub 2016 Jan 22. ACS Nano. 2016. PMID: 26760677 Free PMC article.
-
Multi-modal cryo-EM reveals trimers of protein A10 to form the palisade layer in poxvirus cores.Nat Struct Mol Biol. 2024 Jul;31(7):1114-1123. doi: 10.1038/s41594-023-01201-6. Epub 2024 Feb 5. Nat Struct Mol Biol. 2024. PMID: 38316877 Free PMC article.
-
Lumpy skin disease virus 001/156 protein is a virulence factor that suppresses interferon production through impairing IRF3 dimerization.PLoS Pathog. 2025 Jul 23;21(7):e1013362. doi: 10.1371/journal.ppat.1013362. eCollection 2025 Jul. PLoS Pathog. 2025. PMID: 40700455 Free PMC article.
References
-
- Andres G, Alejo A, Simon-Mateo C, Salas ML. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J.Biol.Chem. 2001;276:780–787. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

