Complete atomistic model of a bacterial cytoplasm for integrating physics, biochemistry, and systems biology
- PMID: 25765281
- PMCID: PMC4388805
- DOI: 10.1016/j.jmgm.2015.02.004
Complete atomistic model of a bacterial cytoplasm for integrating physics, biochemistry, and systems biology
Abstract
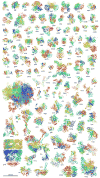
A model for the cytoplasm of Mycoplasma genitalium is presented that integrates data from a variety of sources into a physically and biochemically consistent model. Based on gene annotations, core genes expected to be present in the cytoplasm were determined and a metabolic reaction network was reconstructed. The set of cytoplasmic genes and metabolites from the predicted reactions were assembled into a comprehensive atomistic model consisting of proteins with predicted structures, RNA, protein/RNA complexes, metabolites, ions, and solvent. The resulting model bridges between atomistic and cellular scales, between physical and biochemical aspects, and between structural and systems views of cellular systems and is meant as a starting point for a variety of simulation studies.
Keywords: Crowding; Metabolic reaction network; Mycoplasma genitalium; Protein structure prediction.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



References
-
- Wilhelm BG, Mandad S, Truckenbrodt S, Krohnert K, Schafer C, Rammner B, et al. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science. 2014;344:1023–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

