Characterizing excited conformational states of RNA by NMR spectroscopy
- PMID: 25765780
- PMCID: PMC4755334
- DOI: 10.1016/j.sbi.2015.02.011
Characterizing excited conformational states of RNA by NMR spectroscopy
Abstract
Conformational dynamics is a hallmark of diverse non-coding RNA functions. During these functional processes, RNA molecules almost ubiquitously undergo conformational transitions that are tuned to meet distinct structural and kinetic requirements for proper function. A complete mechanistic understanding of RNA function requires comprehensive structural and dynamic knowledge of these complex transitions, which often involve alternative higher-energy conformational states that pose a major challenge for high-resolution structural study by conventional methods. In this review, we describe recent progress in RNA NMR that has started to unveil detailed structural, thermodynamic and kinetic insights into some of these excited conformational states of RNA and their functional roles in biology.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
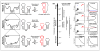
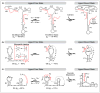

References
-
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. - PubMed
-
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. - PubMed
-
- Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316–323. - PubMed
-
- Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. - PubMed
-
- Williamson JR. Induced fit in RNA–protein recognition. Nat Struct Biol. 2000;7:834–837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

