H2A.Z marks antisense promoters and has positive effects on antisense transcript levels in budding yeast
- PMID: 25765960
- PMCID: PMC4337092
- DOI: 10.1186/s12864-015-1247-4
H2A.Z marks antisense promoters and has positive effects on antisense transcript levels in budding yeast
Abstract
Background: The histone variant H2A.Z, which has been reported to have both activating and repressive effects on gene expression, is known to occupy nucleosomes at the 5' ends of protein-coding genes.
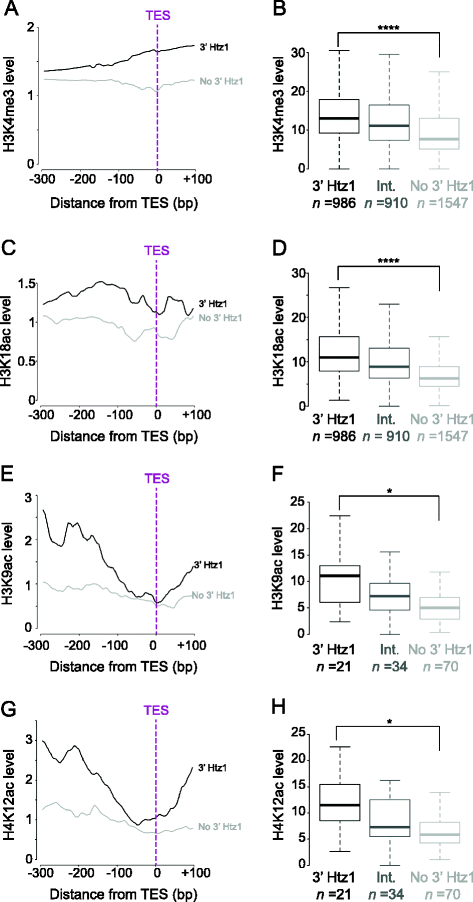
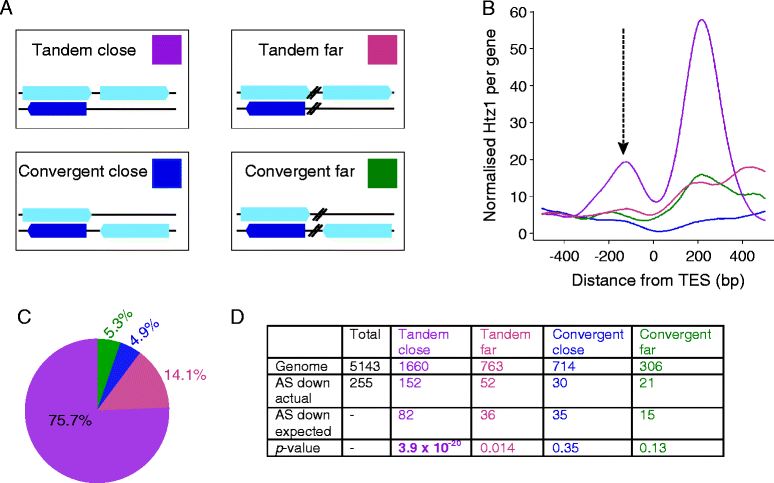
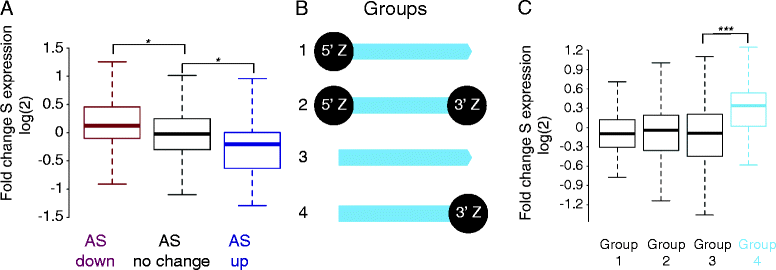
Results: We now find that H2A.Z is also significantly enriched in gene coding regions and at the 3' ends of genes in budding yeast, where it co-localises with histone marks associated with active promoters. By comparing H2A.Z binding to global gene expression in budding yeast strains engineered so that normally unstable transcripts are abundant, we show that H2A.Z is required for normal levels of antisense transcripts as well as sense ones. High levels of H2A.Z at antisense promoters are associated with decreased antisense transcript levels when H2A.Z is deleted, indicating that H2A.Z has an activating effect on antisense transcripts. Decreases in antisense transcripts affected by H2A.Z are accompanied by increased levels of paired sense transcripts.
Conclusions: The effect of H2A.Z on protein coding gene expression is a reflection of its importance for normal levels of both sense and antisense transcripts.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

