Cellular 5'-3' mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses
- PMID: 25766294
- PMCID: PMC4826345
- DOI: 10.1016/j.chom.2015.02.003
Cellular 5'-3' mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses
Abstract
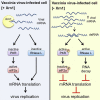
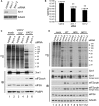
By accelerating global mRNA decay, many viruses impair host protein synthesis, limiting host defenses and stimulating virus mRNA translation. Vaccinia virus (VacV) encodes two decapping enzymes (D9, D10) that remove protective 5' caps on mRNAs, presumably generating substrates for degradation by the host exonuclease Xrn1. Surprisingly, we find VacV infection of Xrn1-depleted cells inhibits protein synthesis, compromising virus growth. These effects are aggravated by D9 deficiency and dependent upon a virus transcription factor required for intermediate and late mRNA biogenesis. Considerable double-stranded RNA (dsRNA) accumulation in Xrn1-depleted cells is accompanied by activation of host dsRNA-responsive defenses controlled by PKR and 2'-5' oligoadenylate synthetase (OAS), which respectively inactivate the translation initiation factor eIF2 and stimulate RNA cleavage by RNase L. This proceeds despite VacV-encoded PKR and RNase L antagonists being present. Moreover, Xrn1 depletion sensitizes uninfected cells to dsRNA treatment. Thus, Xrn1 is a cellular factor regulating dsRNA accumulation and dsRNA-responsive innate immune effectors.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures






Comment in
-
Caps off to poxviruses.Cell Host Microbe. 2015 Mar 11;17(3):287-289. doi: 10.1016/j.chom.2015.02.013. Cell Host Microbe. 2015. PMID: 25766288
References
-
- Aloni Y., Locker H. Symmetrical in vivo transcription of polyoma DNA and the separation of self-complementary viral and cell RNA. Virology. 1973;54:495–505. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

