Transcriptome sequencing reveals potential mechanism of cryptic 3' splice site selection in SF3B1-mutated cancers
- PMID: 25768983
- PMCID: PMC4358997
- DOI: 10.1371/journal.pcbi.1004105
Transcriptome sequencing reveals potential mechanism of cryptic 3' splice site selection in SF3B1-mutated cancers
Abstract
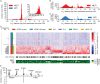
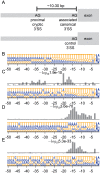
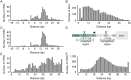
Mutations in the splicing factor SF3B1 are found in several cancer types and have been associated with various splicing defects. Using transcriptome sequencing data from chronic lymphocytic leukemia, breast cancer and uveal melanoma tumor samples, we show that hundreds of cryptic 3' splice sites (3'SSs) are used in cancers with SF3B1 mutations. We define the necessary sequence context for the observed cryptic 3' SSs and propose that cryptic 3'SS selection is a result of SF3B1 mutations causing a shift in the sterically protected region downstream of the branch point. While most cryptic 3'SSs are present at low frequency (<10%) relative to nearby canonical 3'SSs, we identified ten genes that preferred out-of-frame cryptic 3'SSs. We show that cancers with mutations in the SF3B1 HEAT 5-9 repeats use cryptic 3'SSs downstream of the branch point and provide both a mechanistic model consistent with published experimental data and affected targets that will guide further research into the oncogenic effects of SF3B1 mutation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

