Taxonomic Identification of Ruminal Epithelial Bacterial Diversity during Rumen Development in Goats
- PMID: 25769827
- PMCID: PMC4407235
- DOI: 10.1128/AEM.00203-15
Taxonomic Identification of Ruminal Epithelial Bacterial Diversity during Rumen Development in Goats
Abstract
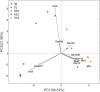
Understanding of the colonization process of epithelial bacteria attached to the rumen tissue during rumen development is very limited. Ruminal epithelial bacterial colonization is of great significance for the relationship between the microbiota and the host and can influence the early development and health of the host. MiSeq sequencing of 16S rRNA genes and quantitative real-time PCR (qPCR) were applied to characterize ruminal epithelial bacterial diversity during rumen development in this study. Seventeen goat kids were selected to reflect the no-rumination (0 and 7 days), transition (28 and 42 days), and rumination (70 days) phases of animal development. Alpha diversity indices (operational taxonomic unit [OTU] numbers, Chao estimate, and Shannon index) increased (P < 0.01) with age, and principal coordinate analysis (PCoA) revealed that the samples clustered together according to age group. Phylogenetic analysis revealed that Proteobacteria, Firmicutes, and Bacteroidetes were detected as the dominant phyla regardless of the age group, and the abundance of Proteobacteria declined quadratically with age (P < 0.001), while the abundances of Bacteroidetes (P = 0.088) and Firmicutes (P = 0.009) increased with age. At the genus level, Escherichia (80.79%) dominated at day zero, while Prevotella, Butyrivibrio, and Campylobacter surged (linearly; P < 0.01) in abundance at 42 and 70 days. qPCR showed that the total copy number of epithelial bacteria increased linearly (P = 0.013) with age. In addition, the abundances of the genera Butyrivibrio, Campylobacter, and Desulfobulbus were positively correlated with rumen weight, rumen papilla length, ruminal ammonia and total volatile fatty acid concentrations, and activities of carboxymethylcellulase (CMCase) and xylanase. Taking the data together, colonization by ruminal epithelial bacteria is age related (achieved at 2 months) and might participate in the anatomic and functional development of the rumen.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



References
-
- Cheng KJ, McCowan RP, Costerton JW. 1979. Adherent epithelial bacteria in ruminants and their roles in digestive-tract function. Am J Clin Nutr 32:139–148. - PubMed
-
- Cho SJ, Cho KM, Shin EC, Lim WJ, Hong SY, Choi BR, Kang JM, Lee SM, Kim YH, Kim H, Yun HD. 2006. 16S rDNA analysis of bacterial diversity in three fractions of cow rumen. J Microbiol Biotechnol 16:92–101.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

