Site history and edaphic features override the influence of plant species on microbial communities in restored tidal freshwater wetlands
- PMID: 25769832
- PMCID: PMC4407224
- DOI: 10.1128/AEM.00038-15
Site history and edaphic features override the influence of plant species on microbial communities in restored tidal freshwater wetlands
Abstract
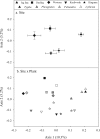
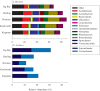
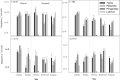
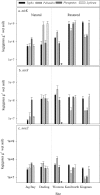
Restored wetland soils differ significantly in physical and chemical properties from their natural counterparts even when plant community compositions are similar, but effects of restoration on microbial community composition and function are not well understood. Here, we investigate plant-microbe relationships in restored and natural tidal freshwater wetlands from two subestuaries of the Chesapeake Bay. Soil samples were collected from the root zone of Typha latifolia, Phragmites australis, Peltandra virginica, and Lythrum salicaria. Soil microbial composition was assessed using 454 pyrosequencing, and genes representing bacteria, archaea, denitrification, methanogenesis, and methane oxidation were quantified. Our analysis revealed variation in some functional gene copy numbers between plant species within sites, but intersite comparisons did not reveal consistent plant-microbe trends. We observed more microbial variations between plant species in natural wetlands, where plants have been established for a long period of time. In the largest natural wetland site, sequences putatively matching methanogens accounted for ∼17% of all sequences, and the same wetland had the highest numbers of genes coding for methane coenzyme A reductase (mcrA). Sequences putatively matching aerobic methanotrophic bacteria and anaerobic methane-oxidizing archaea (ANME) were detected in all sites, suggesting that both aerobic and anaerobic methane oxidation are possible in these systems. Our data suggest that site history and edaphic features override the influence of plant species on microbial communities in restored wetlands.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Bannert A, Kleineidam K, Wissing L, Mueller-Niggemann C, Vogelsang V, Welzl G, Cao Z, Schloter M. 2011. Changes in diversity and functional gene abundances of microbial communities involved in nitrogen fixation, nitrification, and denitrification in a tidal wetland versus paddy soils cultivated for different time periods. J Appl Environ Microbiol 77:6109–6116. doi:10.1128/AEM.01751-10. - DOI - PMC - PubMed
-
- Naeem S, Wright JP. 2003. Disentangling biodiversity effects on ecosystem functioning: deriving solutions to a seemingly insurmountable problem. Ecol Lett 6:567–579. doi:10.1046/j.1461-0248.2003.00471.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

