Species richness and adaptation of marine fungi from deep-subseafloor sediments
- PMID: 25769836
- PMCID: PMC4407237
- DOI: 10.1128/AEM.04064-14
Species richness and adaptation of marine fungi from deep-subseafloor sediments
Abstract
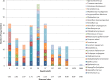
The fungal kingdom is replete with unique adaptive capacities that allow fungi to colonize a wide variety of habitats, ranging from marine habitats to freshwater and terrestrial habitats. The diversity, importance, and ecological roles of marine fungi have recently been highlighted in deep-subsurface sediments using molecular methods. Fungi in the deep-marine subsurface may be specifically adapted to life in the deep biosphere, but this can be demonstrated only using culture-based analyses. In this study, we investigated culturable fungal communities from a record-depth sediment core sampled from the Canterbury Basin (New Zealand) with the aim to reveal endemic or ubiquist adapted isolates playing a significant ecological role(s). About 200 filamentous fungi (68%) and yeasts (32%) were isolated. Fungal isolates were affiliated with the phyla Ascomycota and Basidiomycota, including 21 genera. Screening for genes involved in secondary metabolite synthesis also revealed their bioactive compound synthesis potential. Our results provide evidence that deep-subsurface fungal communities are able to survive, adapt, grow, and interact with other microbial communities and highlight that the deep-sediment habitat is another ecological niche for fungi.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



References
-
- Damare S, Raghukumar C, Raghukumar S. 2006. Fungi in deep-sea sediments of the Central Indian Basin. Deep Sea Res Part I Oceanogr Res Pap 53:14–27. doi:10.1016/j.dsr.2005.09.005. - DOI
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

