Overview of proteomics studies in obstructive sleep apnea
- PMID: 25770042
- PMCID: PMC4402077
- DOI: 10.1016/j.sleep.2014.11.014
Overview of proteomics studies in obstructive sleep apnea
Abstract
Obstructive sleep apnea (OSA) is an underdiagnosed common public health concern causing deleterious effects on metabolic and cardiovascular health. Although much has been learned regarding the pathophysiology and consequences of OSA in the past decades, the molecular mechanisms associated with such processes remain poorly defined. The advanced high-throughput proteomics-based technologies have become a fundamental approach for identifying novel disease mediators as potential diagnostic and therapeutic targets for many diseases, including OSA. Here, we briefly review OSA pathophysiology and the technological advances in proteomics and the first results of its application to address critical issues in the OSA field.
Keywords: Biomarkers; Lung; Mass spectrometry; Metabolic disorders; Obstructive sleep apnea; Proteomics.
Copyright © 2014 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors have declared no conflict of interest.
The ICMJE Uniform Disclosure Form for Potential Conflicts of Interest associated with this article can be viewed by clicking on the following link:
Figures

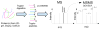
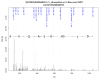
References
-
- Yim S, Jordan A, Malhotra A. Obstructive sleep apnea: clinical presentation, diagnosis and treatment. In: Randerath WJ, Sanner BM, Somers VK, editors. Sleep apnea current diagnosis and treatment. Basel: Karger; 2006. pp. 118–36.
-
- Caples SM, Gami AS, Somers VK. Obstructive sleep apnea. Ann Intern Med. 2005;142:187–97. - PubMed
-
- Pack AI. Advances in sleep-disordered breathing. Am J Respir Crit Care Med. 2006;173:7–15. - PubMed
-
- McNicholas WT, Bonsigore MR Management Committee of ECAB. Sleep apnoea as an independent risk factor for cardiovascular disease: current evidence, basic mechanisms and research priorities. Eur Respir J. 2007;29:156–78. - PubMed
-
- Lurie A. Obstructive sleep apnea in adults: epidemiology, clinical presentation, and treatment options. Adv Cardiol. 2011;46:1–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

