Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato
- PMID: 25770107
- PMCID: PMC4558650
- DOI: 10.1105/tpc.114.132266
Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato
Abstract
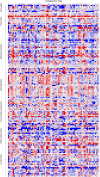
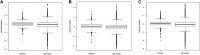
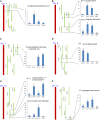
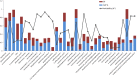
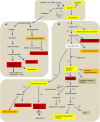
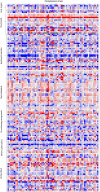
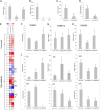
A large-scale metabolic quantitative trait loci (mQTL) analysis was performed on the well-characterized Solanum pennellii introgression lines to investigate the genomic regions associated with secondary metabolism in tomato fruit pericarp. In total, 679 mQTLs were detected across the 76 introgression lines. Heritability analyses revealed that mQTLs of secondary metabolism were less affected by environment than mQTLs of primary metabolism. Network analysis allowed us to assess the interconnectivity of primary and secondary metabolism as well as to compare and contrast their respective associations with morphological traits. Additionally, we applied a recently established real-time quantitative PCR platform to gain insight into transcriptional control mechanisms of a subset of the mQTLs, including those for hydroxycinnamates, acyl-sugar, naringenin chalcone, and a range of glycoalkaloids. Intriguingly, many of these compounds displayed a dominant-negative mode of inheritance, which is contrary to the conventional wisdom that secondary metabolite contents decreased on domestication. We additionally performed an exemplary evaluation of two candidate genes for glycolalkaloid mQTLs via the use of virus-induced gene silencing. The combined data of this study were compared with previous results on primary metabolism obtained from the same material and to other studies of natural variance of secondary metabolism.
© 2015 American Society of Plant Biologists. All rights reserved.
Figures





References
-
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., Angeles E.R., Qian Q., Kitano H., Matsuoka M. (2005). Cytokinin oxidase regulates rice grain production. Science 309: 741–745. - PubMed
-
- Bagheri H., El-Soda M., van Oorschot I., Hanhart C., Bonnema G., Jansen-van den Bosch T., Mank R., Keurentjes J.J.B., Meng L., Wu J., Koornneef M., Aarts M.G.M. (2012). Genetic analysis of morphological traits in a new, versatile, rapid-cycling Brassica rapa recombinant inbred line population. Front. Plant Sci. 3: 183. - PMC - PubMed
-
- Bates, D., Maechler, M., Bolker, B., Walker, S. (2014). lme4: Linear mixed-effects models using Eigen and S4. R package version 1.1-7, http://CRAN.R-project.org/package=lme4.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

