Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt)
- PMID: 25773600
- PMCID: PMC4385436
- DOI: 10.1016/j.molcel.2015.02.008
Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt)
Abstract
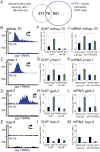
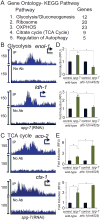
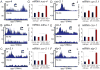
Mitochondrial diseases and aging are associated with defects in the oxidative phosphorylation machinery (OXPHOS), which are the only complexes composed of proteins encoded by separate genomes. To better understand genome coordination and OXPHOS recovery during mitochondrial dysfunction, we examined ATFS-1, a transcription factor that regulates mitochondria-to-nuclear communication during the mitochondrial UPR, via ChIP-sequencing. Surprisingly, in addition to regulating mitochondrial chaperone, OXPHOS complex assembly factor, and glycolysis genes, ATFS-1 bound directly to OXPHOS gene promoters in both the nuclear and mitochondrial genomes. Interestingly, atfs-1 was required to limit the accumulation of OXPHOS transcripts during mitochondrial stress, which required accumulation of ATFS-1 in the nucleus and mitochondria. Because balanced ATFS-1 accumulation promoted OXPHOS complex assembly and function, our data suggest that ATFS-1 stimulates respiratory recovery by fine-tuning OXPHOS expression to match the capacity of the suboptimal protein-folding environment in stressed mitochondria, while simultaneously increasing proteostasis capacity.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


References
-
- Braeckman BP, Houthoofd K, Vanfleteren JR. Assessing metabolic activity in aging Caenorhabditis elegans: concepts and controversies. Aging cell. 2002;1:82–88. discussion 102-103. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

