Beyond Mutations: Additional Mechanisms and Implications of SWI/SNF Complex Inactivation
- PMID: 25774356
- PMCID: PMC4343012
- DOI: 10.3389/fonc.2014.00372
Beyond Mutations: Additional Mechanisms and Implications of SWI/SNF Complex Inactivation
Abstract
SWI/SNF is a major regulator of gene expression. Its role is to facilitate the shifting and exposure of DNA segments within the promoter and other key domains to transcription factors and other essential cellular proteins. This complex interacts with a wide range of proteins and does not function within a single, specific pathway; thus, it is involved in a multitude of cellular processes, including DNA repair, differentiation, development, cell adhesion, and growth control. Given SWI/SNF's prominent role in these processes, many of which are important for blocking cancer development, it is not surprising that the SWI/SNF complex is targeted during cancer initiation and progression both by mutations and by non-mutational mechanisms. Currently, the understanding of the types of alterations, their frequency, and their impact on the SWI/SNF subunits is an area of intense research that has been bolstered by a recent cadre of NextGen sequencing studies. These studies have revealed mutations in SWI/SNF subunits, indicating that this complex is thus important for cancer development. The purpose of this review is to put into perspective the role of mutations versus other mechanisms in the silencing of SWI/SNF subunits, in particular, BRG1 and BRM. In addition, this review explores the recent development of synthetic lethality and how it applies to this complex, as well as how BRM polymorphisms are becoming recognized as potential clinical biomarkers for cancer risk.
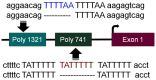
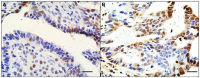
Significance: Recent reviews have detailed the occurrence of mutations in nearly all SWI/SNF subunits, which indicates that this complex is an important target for cancer. However, when the frequency of mutations in a given tumor type is compared to the frequency of subunit loss, it becomes clear that other non-mutational mechanisms must play a role in the inactivation of SWI/SNF subunits. Such data indicate that epigenetic mechanisms that are known to regulate BRM may also be involved in the loss of expression of other SWI/SNF subunits. This is important since epigenetically silenced genes are inducible, and thus, the reversal of the silencing of these non-mutationally suppressed subunits may be a viable mode of targeted therapy.
Keywords: Brahma; SMARCA2; SMARCA4; chromatin remodeling; epigenetic BRG1.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

