Resetting the transcription factor network reverses terminal chronic hepatic failure
- PMID: 25774505
- PMCID: PMC4396487
- DOI: 10.1172/JCI73137
Resetting the transcription factor network reverses terminal chronic hepatic failure
Abstract
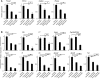
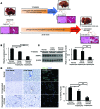
The cause of organ failure is enigmatic for many degenerative diseases, including end-stage liver disease. Here, using a CCl4-induced rat model of irreversible and fatal hepatic failure, which also exhibits terminal changes in the extracellular matrix, we demonstrated that chronic injury stably reprograms the critical balance of transcription factors and that diseased and dedifferentiated cells can be returned to normal function by re-expression of critical transcription factors, a process similar to the type of reprogramming that induces somatic cells to become pluripotent or to change their cell lineage. Forced re-expression of the transcription factor HNF4α induced expression of the other hepatocyte-expressed transcription factors; restored functionality in terminally diseased hepatocytes isolated from CCl4-treated rats; and rapidly reversed fatal liver failure in CCl4-treated animals by restoring diseased hepatocytes rather than replacing them with new hepatocytes or stem cells. Together, the results of our study indicate that disruption of the transcription factor network and cellular dedifferentiation likely mediate terminal liver failure and suggest reinstatement of this network has therapeutic potential for correcting organ failure without cell replacement.
Figures




Comment in
-
Liver: Reprogramming a hepatocyte's memory of liver disease.Nat Rev Gastroenterol Hepatol. 2015 May;12(5):250. doi: 10.1038/nrgastro.2015.51. Epub 2015 Mar 31. Nat Rev Gastroenterol Hepatol. 2015. PMID: 25824999 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

