Susceptibility to tuberculosis is associated with variants in the ASAP1 gene encoding a regulator of dendritic cell migration
- PMID: 25774636
- PMCID: PMC4414475
- DOI: 10.1038/ng.3248
Susceptibility to tuberculosis is associated with variants in the ASAP1 gene encoding a regulator of dendritic cell migration
Abstract
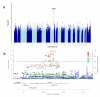
Human genetic factors predispose to tuberculosis (TB). We studied 7.6 million genetic variants in 5,530 people with pulmonary TB and in 5,607 healthy controls. In the combined analysis of these subjects and the follow-up cohort (15,087 TB patients and controls altogether), we found an association between TB and variants located in introns of the ASAP1 gene on chromosome 8q24 (P = 2.6 × 10(-11) for rs4733781; P = 1.0 × 10(-10) for rs10956514). Dendritic cells (DCs) showed high ASAP1 expression that was reduced after Mycobacterium tuberculosis infection, and rs10956514 was associated with the level of reduction of ASAP1 expression. The ASAP1 protein is involved in actin and membrane remodeling and has been associated with podosomes. The ASAP1-depleted DCs showed impaired matrix degradation and migration. Therefore, genetically determined excessive reduction of ASAP1 expression in M. tuberculosis-infected DCs may lead to their impaired migration, suggesting a potential mechanism of predisposition to TB.
Figures


Comment in
-
Regulator of dendritic cell migration, ASAP1 is associated with increased susceptibility to tuberculosis.Clin Genet. 2015 Dec;88(6):530-1. doi: 10.1111/cge.12611. Epub 2015 Jun 15. Clin Genet. 2015. PMID: 25970765
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

