SYBR Green real-time PCR-RFLP assay targeting the plasmodium cytochrome B gene--a highly sensitive molecular tool for malaria parasite detection and species determination
- PMID: 25774805
- PMCID: PMC4361616
- DOI: 10.1371/journal.pone.0120210
SYBR Green real-time PCR-RFLP assay targeting the plasmodium cytochrome B gene--a highly sensitive molecular tool for malaria parasite detection and species determination
Abstract
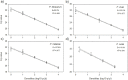
A prerequisite for reliable detection of low-density Plasmodium infections in malaria pre-elimination settings is the availability of ultra-sensitive and high-throughput molecular tools. We developed a SYBR Green real-time PCR restriction fragment length polymorphism assay (cytb-qPCR) targeting the cytochrome b gene of the four major human Plasmodium species (P. falciparum, P. vivax, P. malariae, and P. ovale) for parasite detection and species determination with DNA extracted from dried blood spots collected on filter paper. The performance of cytb-qPCR was first compared against four reference PCR methods using serially diluted Plasmodium samples. The detection limit of the cytb-qPCR was 1 parasite/μl (p/μl) for P. falciparum and P. ovale, and 2 p/μl for P. vivax and P. malariae, while the reference PCRs had detection limits of 0.5-10 p/μl. The ability of the PCR methods to detect low-density Plasmodium infections was then assessed using 2977 filter paper samples collected during a cross-sectional survey in Zanzibar, a malaria pre-elimination setting in sub-Saharan Africa. Field samples were defined as 'final positive' if positive in at least two of the five PCR methods. Cytb-qPCR preformed equal to or better than the reference PCRs with a sensitivity of 100% (65/65; 95%CI 94.5-100%) and a specificity of 99.9% (2910/2912; 95%CI 99.7-100%) when compared against 'final positive' samples. The results indicate that the cytb-qPCR may represent an opportunity for improved molecular surveillance of low-density Plasmodium infections in malaria pre-elimination settings.
Conflict of interest statement
Figures




Similar articles
-
Human malaria diagnosis using a single-step direct-PCR based on the Plasmodium cytochrome oxidase III gene.Malar J. 2016 Feb 29;15:128. doi: 10.1186/s12936-016-1185-x. Malar J. 2016. PMID: 26928594 Free PMC article.
-
Towards high-throughput molecular detection of Plasmodium: new approaches and molecular markers.Malar J. 2009 Apr 29;8:86. doi: 10.1186/1475-2875-8-86. Malar J. 2009. PMID: 19402894 Free PMC article.
-
Sub-microscopic malaria cases and mixed malaria infection in a remote area of high malaria endemicity in Rattanakiri province, Cambodia: implication for malaria elimination.Malar J. 2010 Apr 22;9:108. doi: 10.1186/1475-2875-9-108. Malar J. 2010. PMID: 20409349 Free PMC article.
-
Two sympatric types of Plasmodium ovale and discrimination by molecular methods.J Microbiol Immunol Infect. 2017 Oct;50(5):559-564. doi: 10.1016/j.jmii.2016.08.004. Epub 2016 Dec 18. J Microbiol Immunol Infect. 2017. PMID: 28065415 Review.
-
Malaria rapid diagnostic tests in elimination settings--can they find the last parasite?Clin Microbiol Infect. 2011 Nov;17(11):1624-31. doi: 10.1111/j.1469-0691.2011.03639.x. Epub 2011 Sep 13. Clin Microbiol Infect. 2011. PMID: 21910780 Free PMC article. Review.
Cited by
-
Detection of Plasmodium falciparum by Light Microscopy, Loop-Mediated Isothermal Amplification, and Polymerase Chain Reaction on Day 3 after Initiation of Artemether-Lumefantrine Treatment for Uncomplicated Malaria in Bagamoyo District, Tanzania: A Comparative Trial.Am J Trop Med Hyg. 2019 Nov;101(5):1144-1147. doi: 10.4269/ajtmh.19-0298. Am J Trop Med Hyg. 2019. PMID: 31549618 Free PMC article.
-
Significant number of Plasmodium vivax mono-infections by PCR misidentified as mixed infections (P. vivax/P. falciparum) by microscopy and rapid diagnostic tests: malaria diagnostic challenges in Ethiopia.Malar J. 2023 Jul 1;22(1):201. doi: 10.1186/s12936-023-04635-x. Malar J. 2023. PMID: 37393257 Free PMC article.
-
Human malaria diagnosis using a single-step direct-PCR based on the Plasmodium cytochrome oxidase III gene.Malar J. 2016 Feb 29;15:128. doi: 10.1186/s12936-016-1185-x. Malar J. 2016. PMID: 26928594 Free PMC article.
-
Prevalence and Characteristics of Plasmodium vivax Gametocytes in Duffy-Positive and Duffy-Negative Populations across Ethiopia.Am J Trop Med Hyg. 2024 Apr 16;110(6):1091-1099. doi: 10.4269/ajtmh.23-0877. Print 2024 Jun 5. Am J Trop Med Hyg. 2024. PMID: 38626749 Free PMC article.
-
Performance of rapid diagnostic test, blood-film microscopy and PCR for the diagnosis of malaria infection among febrile children from Korogwe District, Tanzania.Malar J. 2016 Jul 26;15(1):391. doi: 10.1186/s12936-016-1450-z. Malar J. 2016. PMID: 27459856 Free PMC article.
References
-
- WHO World malaria report 2013 Geneva: World Health Organization; 2013.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

