RECQ1 interacts with FEN-1 and promotes binding of FEN-1 to telomeric chromatin
- PMID: 25774876
- PMCID: PMC4441847
- DOI: 10.1042/BJ20141021
RECQ1 interacts with FEN-1 and promotes binding of FEN-1 to telomeric chromatin
Abstract
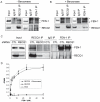
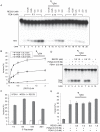
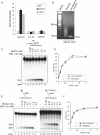
RecQ helicases are a family of highly conserved proteins that maintain genomic stability through their important roles in replication restart mechanisms. Cellular phenotypes of RECQ1 deficiency are indicative of aberrant repair of stalled replication forks, but the molecular functions of RECQ1, the most abundant of the five known human RecQ homologues, have remained poorly understood. We show that RECQ1 associates with FEN-1 (flap endonuclease-1) in nuclear extracts and exhibits direct protein interaction in vitro. Recombinant RECQ1 significantly stimulated FEN-1 endonucleolytic cleavage of 5'-flap DNA substrates containing non-telomeric or telomeric repeat sequence. RECQ1 and FEN-1 were constitutively present at telomeres and their binding to the telomeric chromatin was enhanced following DNA damage. Telomere residence of FEN-1 was dependent on RECQ1 since depletion of RECQ1 reduced FEN-1 binding to telomeres in unperturbed cycling cells. Our results confirm a conserved collaboration of human RecQ helicases with FEN-1 and suggest both overlapping and specialized roles of RECQ1 in the processing of DNA structure intermediates proposed to arise during replication, repair and recombination.
Figures






References
-
- Larsen N, Hickson I. RecQ Helicases: Conserved Guardians of Genomic Integrity. In: Spies M, editor. In DNA Helicases and DNA Motor Proteins. Springer; New York: 2013. pp. 161–184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

