Identification of protein tyrosine phosphatase receptor gamma extracellular domain (sPTPRG) as a natural soluble protein in plasma
- PMID: 25775014
- PMCID: PMC4361625
- DOI: 10.1371/journal.pone.0119110
Identification of protein tyrosine phosphatase receptor gamma extracellular domain (sPTPRG) as a natural soluble protein in plasma
Abstract
Background: PTPRG is a widely expressed protein tyrosine phosphatase present in various isoforms. Peptides from its extracellular domain have been detected in plasma by proteomic techniques. We aim at characterizing the plasmatic PTPRG (sPTPRG) form and to identify its source.
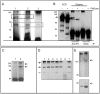
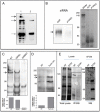
Methodology/principal findings: The expression of sPTPRG was evaluated in human plasma and murine plasma and tissues by immunoprecipitation and Western blotting. The polypeptides identified have an apparent Mr of about 120 kDa (major band) and 90 kDa (minor band) respectively. Full length PTPRG was identified in the 100.000×g pelleted plasma fraction, suggesting that it was present associated to cell-derived vesicles (exosomes). The release of sPTPRG by HepG2 human hepatocellular carcinoma cell line was induced by ethanol and sensitive to metalloproteinase and not to Furin inhibitors. Finally, increased levels of the plasmatic ∼120 kDa isoform were associated with the occurrence of liver damage.
Conclusions: These results demonstrate that sPTPRG represent a novel candidate protein biomarker in plasma whose increased expression is associated to hepatocyte damage. This observation could open a new avenue of investigation in this challenging field.
Conflict of interest statement
Figures




References
-
- Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004; 304: 1164–1166. - PubMed
-
- Panagopoulos I, Pandis N, Thelin S, Petersson C, Mertens F, Borg A, et al. The FHIT and PTPRG genes are deleted in benign proliferative breast disease associated with familial breast cancer and cytogenetic rearrangements of chromosome band 3p14. Cancer Res. 1996; 56: 4871–4875. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

